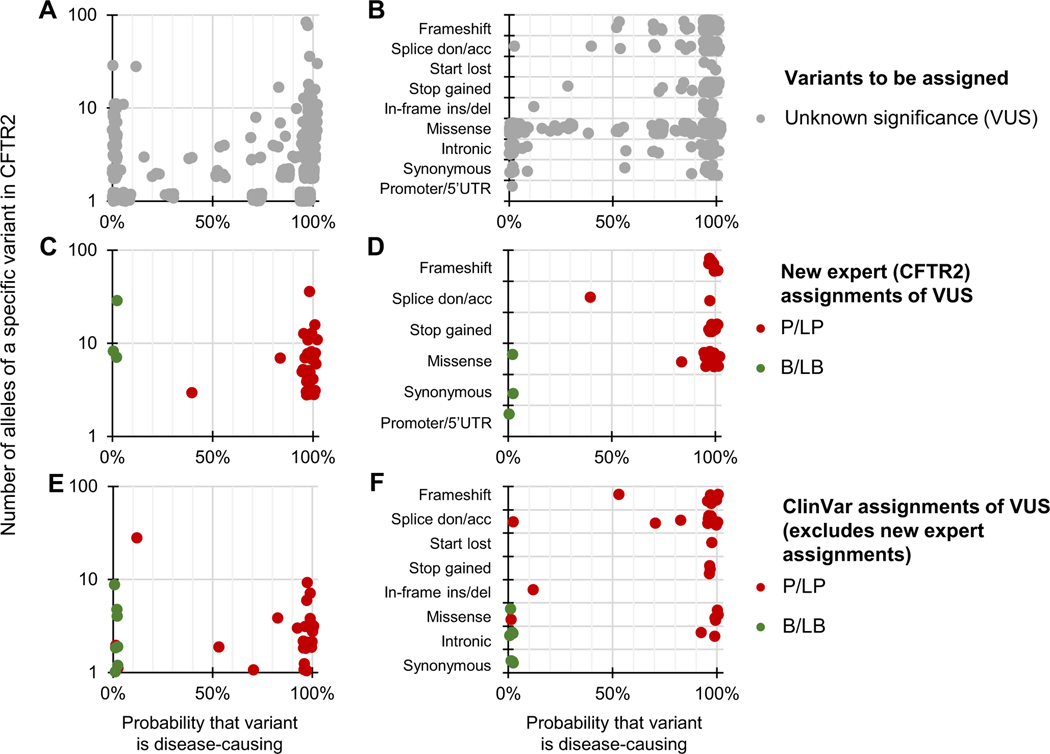
Figure 2. Differentiating CFTR variants of unknown significance with Bayesian prevalence ratio (BayPR).
A-F. Data are jittered for display purposes only; precise allele counts and BayPR probabilities are provided in Supplemental Table 1. A. CFTR variants that had not undergone interpretation by CFTR2 (n = 969; gray dots) cluster primarily into high (>90%; 735 variants) or low (<10%; 129 variants) probabilities. B. BayPR probabilities of unassigned CFTR variants stratified according to their functional consequence. C. A total of 52 unassigned CFTR variants were subsequently and independently assessed for disease liability by CFTR2 using clinical and functional criteria. D. BayPR probabilities of newly-assigned CFTR variants stratified according to their functional consequence. E. CFTR variants that remain without a CFTR2 interpretation and which have P/LP or B/LB interpretations in ClinVar (n = 41) cluster primarily into high (>90%; 28 variants) or low (<10%; 9 variants) probabilities. F. BayPR probabilities of ClinVar-assigned P/LP or B/LB CFTR variants without a CFTR2 annotation parsed by predicted functional effect. acc, acceptor; B, benign; CFTR2, Clinical and Functional Translation of CFTR; del, deletion; don, donor; ins, insertion; LB, likely benign; LP, likely pathogenic; P, pathogenic; UTR, untranslated region; VUS, variant of unknown significance.