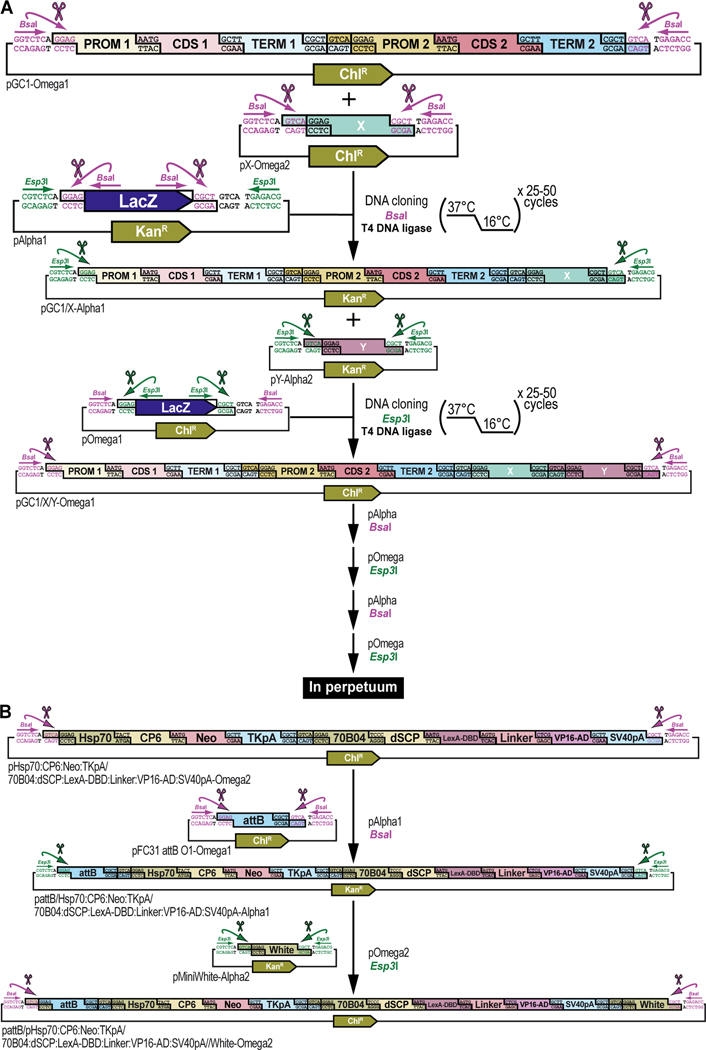
Figure 13. Further assembly steps to expand genetic constructs towards final complexity for a multiplexed selection and counterselection genetic strategy using Drosophila melanogaster.
(A) General principles of further assembly steps using pAlpha and pOmega level vector backbones to expand genetic constructs towards final complexity. Omega level assembly products (see Figure 11 and Figure 12) can be further pairwise expanded into an Alpha level destination vector again using the BsaI cloning enzyme. GoldenBraid2.0 assembly is iterative, always bringing together a pair of vectors of the same level, but not the same cloning identity (e.g., assembling Omega1 and Omega2 together is valid, while assembling Omega1 and Omega1 or Omega2 and Omega2 together is not) using a similar grammar system to ensure ordered assembly of vector inserts (see Figure 2C). Thus, an insert in Omega1 always assembles 5’ of the insert in the accompanying vector, Omega2. Hence, the two transcription units assembled in pOmega1 can be expanded with any assembly product (“X”) located in pOmega2 into one of the Alpha destination vectors. Both plasmid assemblies, each encoding an assembly of various complexity, are combined with an Alpha destination vector (pAlpha1 or pAlpha2), the Type IIs restriction enzyme BsaI, and T4 DNA ligase (together with 10x T4 DNA ligase buffer). The assembly protocol cycles 25 to 50 times between 37°C (favoring cutting using BsaI) and 16°C (favoring ligation using T4 DNA ligase). Assembled plasmids are identified and characterized similarly as described before (see Figure 9 and Figure 10). The resulting product will consist of two transcription units expanded with assembly “X” in the order dictated by their Omega level cloning grammar identities. This assembly product itself can be further pairwise expanded into an Omega level destination vector again using the Esp3I cloning enzyme. Hence, two transcription units expanded with assembly “X” in pAlpha1, can be expanded with any assembly product (“Y”) located in pAlpha2 using one of the Omega destination vectors. Both plasmid assemblies, each encoding an assembly of various complexity, are combined with an Omega destination vector (pOmega1 or pOmega2), the Type IIs restriction enzyme Esp3I, and T4 DNA ligase (together with 10x T4 DNA ligase buffer). The assembly protocol cycles 25 to 50 times between 37°C (favoring cutting) and 16°C (favoring ligation). Assembled plasmids are identified and characterized similarly as described before (see Figure 11 and Figure 12). The resulting product will consist of two transcription units and assembly “X”, expanded with assembly “Y” in the order dictated by their Alpha level cloning grammar identities. Application of the principles described here are illustrated below (see B) for finalizing the G418 sulfate-selectable LexA transactivator plasmid pattB/Hsp70:CP6:Neo:TKpA/70B04:dSCP:LexA-DBD:Linker:VP16-AD:SV40pA/White-Omega2 (see Figure 3). (B) Further assembly steps to finalize the G418 sulfate-selectable LexA transactivator plasmid pattB/Hsp70:CP6:Neo:TKpA/70B04:dSCP:LexA-DBD:Linker:VP16-AD:SV40pA/White-Omega2. Additional assembly steps required to build the G418 sulfate-selectable LexA transactivator plasmid for tissue-specific overexpression within the R70B04 expression domain. The dual transcription unit Hsp70:CP6:Neo:TKpA/70B04:dSCP:LexA-DBD:Linker:VP16-AD:SV40pA in pOmega2 is further expanded with a phiC31 bacteriophage attB attachment site for site-specific transgenesis (attB) in pOmega1 to form pattB/Hsp70:CP6:Neo:TKpA/70B04:dSCP:LexA-DBD:Linker:VP16-AD:SV40pA in pAlpha1, which itself is further expanded with a dominant “eye” screening marker called “mini-white” from Drosophila melanogaster (White) in pAlpha2 to form pattB/Hsp70:CP6:Neo:TKpA/70B04:dSCP:LexA-DBD:Linker:VP16-AD:SV40pA/White in pOmega2 (abbreviated to G418 sulfate-selectable LexA transactivator plasmid) (see Figure 3).