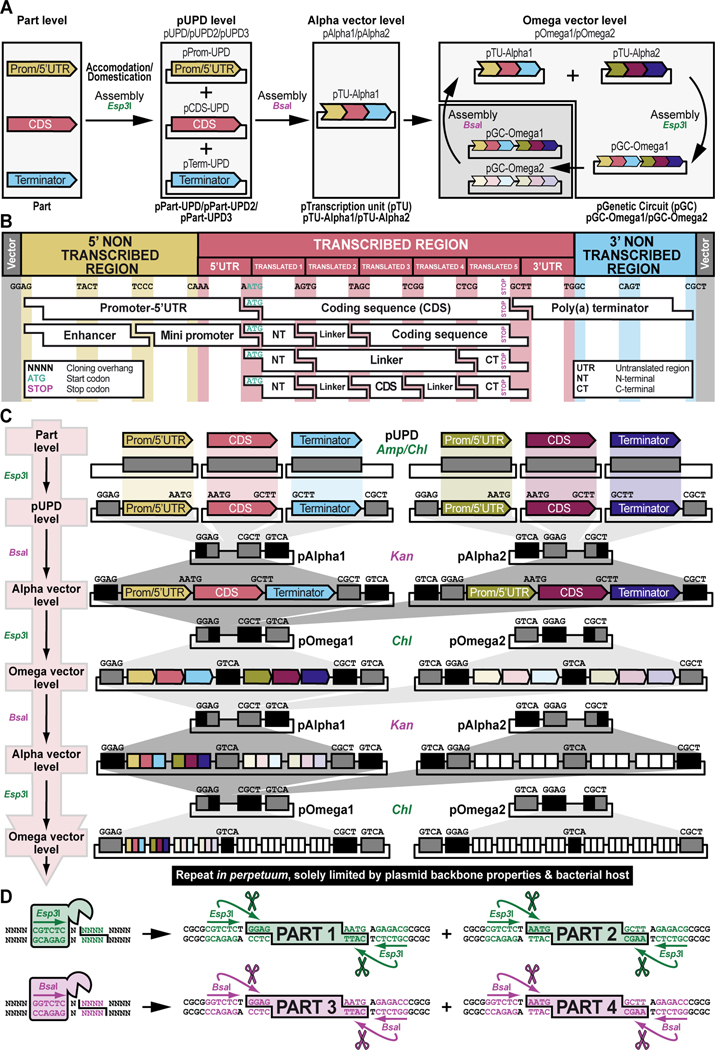
Figure 2. Schematic overview of GoldenBraid 2.0 synthetic assembly DNA cloning to build plasmids for multiplexed selection, counterselection or other genetic strategies using Drosophila melanogaster.
(A) Simplified schematic of the synthetic assembly DNA cloning workflow in GoldenBraid 2.0. DNA parts, including promoter (Prom), coding DNA sequence (CDS), and terminator (Term) (Part level), are first accommodated (also known as domesticated) to the GoldenBraid 2.0 (GB2.0) workflow via synthetic assembly DNA cloning, with the Type IIs restriction enzyme Esp3I, into a Universal Part Domesticator plasmid (pUPD, pUPD2 or pUPD3). This produces a library of DNA part plasmids (pPart-UPD), i.e., pProm-UPD, pCDS-UPD, or pTerm-UPD, respectively (pUPD level), with defined overhangs (see B), allowing ordered assembly in the next assembly level (see C). These parts are further assembled into an Alpha level destination vector (pAlpha1 or pAlpha2) using the Type IIs restriction enzyme BsaI, resulting in a plasmid containing a transcription unit, pTranscription unit (pTU), defined by at least a promoter, coding sequence and terminator (pTU-Alpha1 or pTU-Alpha2), or another assembly of various complexity (Alpha vector level). Finally, Alpha level vectors can be further assembled into an Omega level destination vector (pOmega1 or pOmega2) using again the Type IIs restriction enzyme Esp3I, resulting in a plasmid containing a genetic circuit, pGenetic circuit (pGC), defined by two transcription units or other DNA assemblies of various complexity (pGC-Omega1 or pGC-Omega2) (Omega vector level). Conveniently, those Omega assemblies can be further pairwise assembled into an Alpha level destination vector, and the resulting product can serve as a reagent for further assembly reactions into an Omega level destination vector, and so on. (B) Underlying cloning grammar defined by orthogonal restriction enzyme overhangs to guide ordered synthetic assembly DNA cloning of DNA parts in a multipartite fashion by GoldenBraid 2.0 assembly. GB2.0 cloning, like Golden Gate assembly from which it is derived, features a predefined system of 4-nucleotide cloning overhangs used to assemble linearized DNA parts together in a defined fashion. These overhangs form a “cloning grammar” which governs part identity and assembly order such that a promoter type part always assembles 5’ of a coding sequence which assembles 5’ of a terminator sequence and so on. Several 5’ – 3’ cloning overhangs and their associated part identities are shown to illustrate how different parts can be assembled in meaningful ways. (C) Detailed cloning workflow defined by orthogonal restriction enzyme overhangs to guide parts and intermediate assemblies through the GoldenBraid 2.0 assembly pipeline to reach final status. DNA parts are cloned in Universal Part Domesticator plasmids (pUPD, pUPD2 or pUPD3) using the Type IIs restriction enzyme Esp3I and selected for with ampicillin or chloramphenicol. Cloned parts are combined in a meaningful manner into an Alpha level destination vector (pAlpha1 or pAlpha2) using the Type IIs restriction enzyme BsaI and selected for with kanamycin. Alpha level assemblies can be further pairwise combined into an Omega level destination vector (pOmega1 or pOmega2) using again the Type IIs restriction enzyme Esp3I and selected for with chloramphenicol. Those Omega assemblies can be further pairwise combined into an Alpha level destination vector, and the resulting Alpha assembly can be further pairwise combined into an Omega level destination vector, and so on. This infinitely iterative process always involves a pair of vectors of the same level but different identity (e.g., pAlpha1 and pAlpha2 is a valid assembly into pOmega1 or pOmega2, but pAlpha1 and pAlpha1 is not) governed by their own cloning grammars. This is true for both Alpha and Omega assemblies: pAlpha1 always combines with pAlpha2 in a defined order into any of the two Omega level plasmids, and pOmega1 and pOmega2 always combine in a defined order into any of the two Alpha level plasmids. The ordered assembly of pAlpha1 and pAlpha2 into Omega level vectors is defined by the 5’ “GGAG” and 3’ “GTCA” grammar of pAlpha1 and the 5’ “GTCA” and 3’ “CGCT” grammar of pAlpha2, combined by switching antibiotic selection from kanamycin (for Alpha vectors) to chloramphenicol (for Omega vectors) (see Figure 9 and Figure 10). The same overhangs define the ordered assembly of pOmega1 and pOmega2 into Alpha level vectors, except that antibiotic selection is switched from chloramphenicol (for Omega vectors) to kanamycin (for Alpha vectors) (see Figure 11 and Figure 12). Not all Alpha or Omega assemblies have to be multipart assemblies, as long as the 5’ end has “GGAG” grammar and the 3’ end has “CGCT” grammar. This allows flexibility in the assemblies when a less complex part, e.g., just an attB attachment site, is needed to expand an already complex assembly (see Figure 3 and Figure 13). (D) Simplified schematic of the two Type IIs restriction enzymes used by GoldenBraid 2.0 assembly. Esp3I (Top), or its isoschizomer BsmBI, is used during accommodating/domesticating assemblies going from Part level to pUPD level (see A and C), as well as during assemblies going from Alpha vector level to Omega vector level (see A and C). BsaI (Bottom) is used during assemblies going from pUPD level to Alpha vector level (see A and C), as well as during assemblies going from Omega vector level back to Alpha vector level (see A and C). Both enzymes bind to a unique 6-bp recognition site and create sticky ends one base pair away from their binding site, leaving a 5’ sticky 4-nucleotide sequence behind that can be used for annealing purposes during the ligation step of assembly cloning. Since this 5’ sticky 4-nucleotide sequence is nonspecific (“NNNN”), it can be made user-specific, i.e., programmable, as needed (see B), allowing ordered assemblies to happen using those overhangs, as illustrated for two parts for each enzyme, part 1 and 2 can ligate together using “AATG” sticky ends after cutting with Esp3I, while part 3 and 4 can ligate together using “AATG” sticky ends after cutting with BsaI.