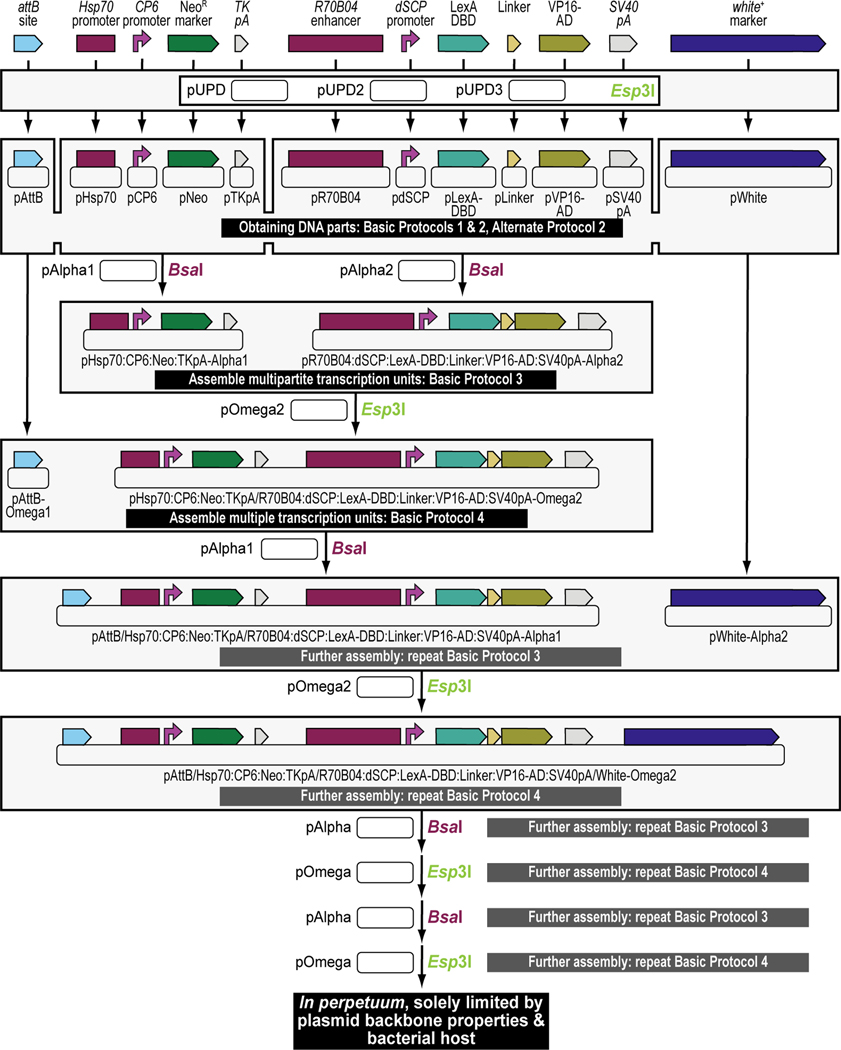
Figure 3. Experimental steps during a typical synthetic assembly DNA cloning workflow to build a genetic construct of continuously increasing complexity: building a G418 sulfate-selectable LexA transactivator plasmid for tissue-specific overexpression as an example.
First, all parts needed for assembly are accommodated or “domesticated” into a Universal Part Domesticator plasmid (pUPD, pUPD2 or pUPD3) using Esp3I, as described in Basic Protocol 1, Basic Protocol 2, and Alternate Protocol 2. Parts include a ФC31 bacteriophage attB attachment site for site-specific transgenesis (attB site), the Hsp70 promoter from Drosophila melanogaster (Hsp70 promoter), the synthetic Escherichia coli CP6 promoter (CP6 promoter), the Neomycin phosphotransferase II of transposon Tn5 (NeoR marker), the minimal polyadenylation signal of the thymidine kinase gene from the herpes simplex virus (TK pA), the R70B04 enhancer from Drosophila melanogaster (R70B04 enhancer), the Drosophila melanogaster synthetic core promoter (dSCP promoter), the DNA binding domain of the LexA repressor from Escherichia coli (LexA DBD), a (GlyGlyGlySer)4 peptide linker (Linker), the transcription factor activation domain of VP16 from the herpes simplex virus (VP16-AD), the late polyadenylation signal from simian vacuolating virus 40 (SV40 pA), and the dominant “eye” screening marker called “mini-white” from Drosophila melanogaster (white+ marker). Next, several of these parts are assembled in Alpha level vector backbones to form transcription units using BsaI, as described in Basic Protocol 3. The Hsp70 promoter, the CP6 promoter, the NeoR marker, and the TK pA are assembled in pAlpha1 resulting in the plasmid pHsp70:CP6:Neo:TKpA-Alpha1, while the R70B04 enhancer, the dSCP promoter, the LexA DBD, Linker, the VP16-AD, and the SV40 pA are assembled in pAlpha2 resulting in the plasmid pR70B04:dSCP:LexA-DBD:Linker:VP16-AD:SV40pA-Alpha2. Alpha assemblies can be further combined in Omega level vector backbones using Esp3I, to form genetic circuits, defined by two transcription units or other DNA assemblies of various complexity, as described in Basic Protocol 4. Alpha assemblies, pHsp70:CP6:Neo:TKpA-Alpha1 and pR70B04:dSCP:LexA-DBD:Linker:VP16-AD:SV40pA-Alpha2 are combined in pOmega2 using Esp3I, resulting in plasmid pHsp70:CP6:Neo:TKpA/R70B04:dSCP:LexA-DBD:Linker:VP16-AD:SV40pA-Omega2. This assembly, together with the attB site located in a pOmega1 plasmid are further combined in pAlpha1 using BsaI, resulting in plasmid pAttB/Hsp70:CP6:Neo:TKpA/R70B04:dSCP:LexA-DBD:Linker:VP16-AD:SV40pA-Alpha1 (repeat Basic Protocol 3), which then, together with the white+ marker located in a pAlpha2 plasmid, is additionally expanded in a pOmega2 plasmid using Esp3I to form the final plasmid, pAttB/Hsp70:CP6:Neo:TKpA/R70B04:dSCP:LexA-DBD:Linker:VP16-AD:SV40pA/White-Omega2, also known as the G418 sulfate-selectable LexA transactivator plasmid for tissue-specific overexpression within the R70B04 expression domain (repeat Basic Protocol 4). If needed, this plasmid can be further expanded using additional rounds of synthetic assembly DNA cloning using BsaI (repeat Basic Protocol 3) or Esp3I (repeat Basic Protocol 4), as indicated. Essentially, assemblies could occur in perpetuum, solely limited by plasmid backbone properties (a high-copy number plasmid backbone can maintain up to about 20 kilobases of insert, while a low-copy number plasmid backbone could maintain several hundreds of kilobases of insert) and the large plasmid maintenance capabilities of the bacterial host (some bacterial hosts cannot maintain large insert plasmids while others can). Combined with an appropriate LexA-Op responder plasmid, e.g., the Blasticidin S-selectable LexA-Op responder plasmid (see Figure 4), both plasmids can then be used to obtain a double transgenic fly through multiplexed dual selection transgenesis (see Venken et al., 2023) that can be used to determine gene expression patterns (See Figure 15).