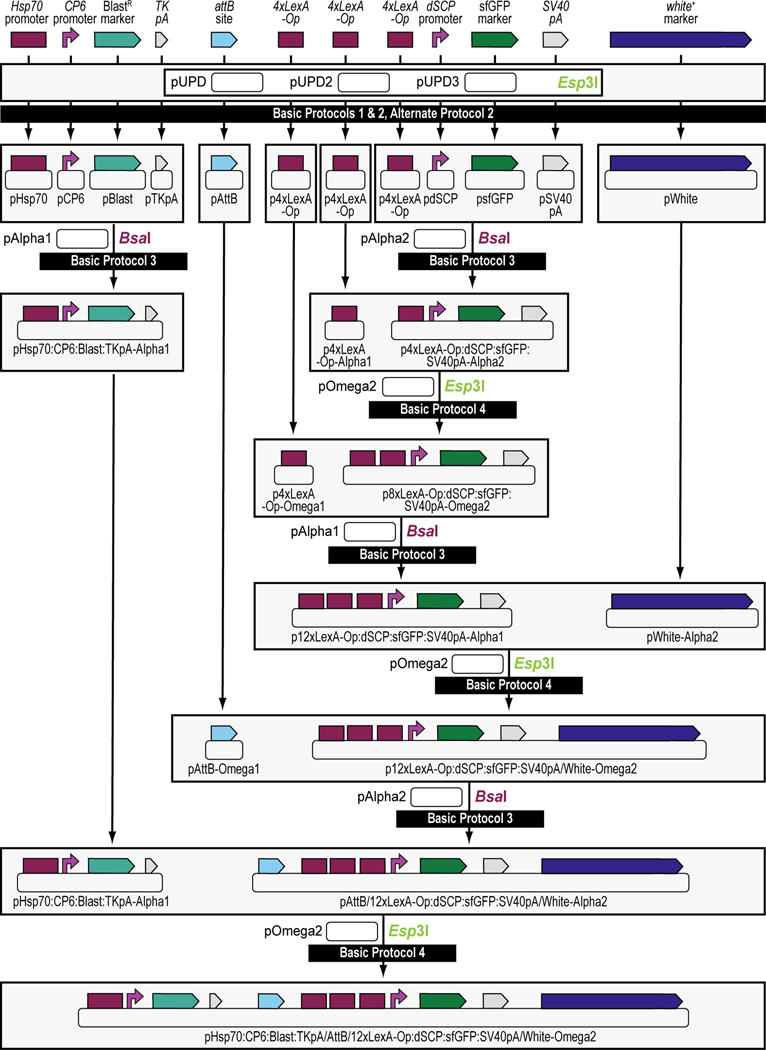
Figure 4. Schematic overview of the experimental steps during the synthetic assembly cloning workflow to build a Blasticidin S-selectable LexA-Op responder plasmid for tissue-specific overexpression in Drosophila melanogaster.
First, all parts needed for assembly are domesticated into a Universal Part Domesticator plasmid (pUPD, pUPD2 or pUPD3) using Esp3I (Basic Protocol 1, Basic Protocol 2, or Alternate Protocol 2). Parts are: the Hsp70 promoter from Drosophila melanogaster (Hsp70 promoter), the synthetic Escherichia coli CP6 promoter (CP6 promoter), the Blasticidin S resistance deaminase gene (BlastR marker), the minimal polyadenylation signal of the thymidine kinase gene from the herpes simplex virus (TK pA), a ФC31 bacteriophage attB attachment site for site-specific transgenesis (attB site), three times 4 copies of the binding site for the LexA DNA binding domain (4xLexA-Op), the Drosophila melanogaster synthetic core promoter (dSCP promoter), the green fluorescent protein reporter sfGFP (sfGFP marker), the late polyadenylation signal from simian vacuolating virus 40 (SV40 pA), and the dominant “eye” screening marker called “mini-white” from Drosophila melanogaster (white+ marker). Next several of these parts are assembled in Alpha level vector backbones to form transcription units using BsaI (see Basic Protocol 3). The Hsp70 promoter, the CP6 promoter, the BlastR marker, and the TK pA are assembled in pAlpha1, resulting in plasmid pHsp70:CP6:Blast:TKpA-Alpha1, while one copy of 4xLexA-Op, the dSCP promoter, the sfGFP marker, and the SV40 pA are assembled together in pAlpha2, resulting in plasmid p4xLexA-Op:dSCP:sfGFP:SV40pA-Alpha2. The latter assembly, together with the second copy of 4xLexA-Op located in pAlpha1 are combined in pOmega2 using Esp3I, resulting in plasmid p8xLexA-Op:dSCP:sfGFP:SV40pA-Alpha2 (see Basic Protocol 4), which together with the third and final copy of 4xLexA-Op located in pOmega1 are combined in pAlpha1 using BsaI, resulting in plasmid p12xLexA-Op:dSCP:sfGFP:SV40pA-Alpha2 (see Basic Protocol 3). This assembly, together with the white+ marker located in a pAlpha2 plasmid, is further combined in a pOmega2 plasmid using Esp3I resulting in plasmid p12xLexA-Op:dSCP:sfGFP:SV40pA/White-Omega2 (see Basic Protocol 4), to which the attB site located in a pOmega1 plasmid is added in pAlpha2 using BsaI, resulting in plasmid pAttB/12xLexA-Op:dSCP:sfGFP:SV40pA/White-Alpha2 (see Basic Protocol 3). During a final step, the Hsp70:CP6:Blast:TKpA-Alpha1 transcription unit located in pAlpha1 and the previous assembly in pAlpha2 are combined together in pOmega2 using Esp3I to form the final plasmid pHsp70:CP6:Blast:TKpA/AttB/12xLexA-Op:dSCP:sfGFP:SV40pA/White-Omega2, also known as the Blasticidin S-selectable LexA-Op responder plasmid (see Basic Protocol 4). Combined with an appropriate LexA transactivator plasmid, e.g., the G418 sulfate-selectable LexA transactivator plasmid (see Figure 3), both plasmids can then be used to obtain a double transgenic fly through multiplexed dual selection transgenesis (see Venken et al., 2023) that can be used to determine gene expression patterns (See Figure 15).