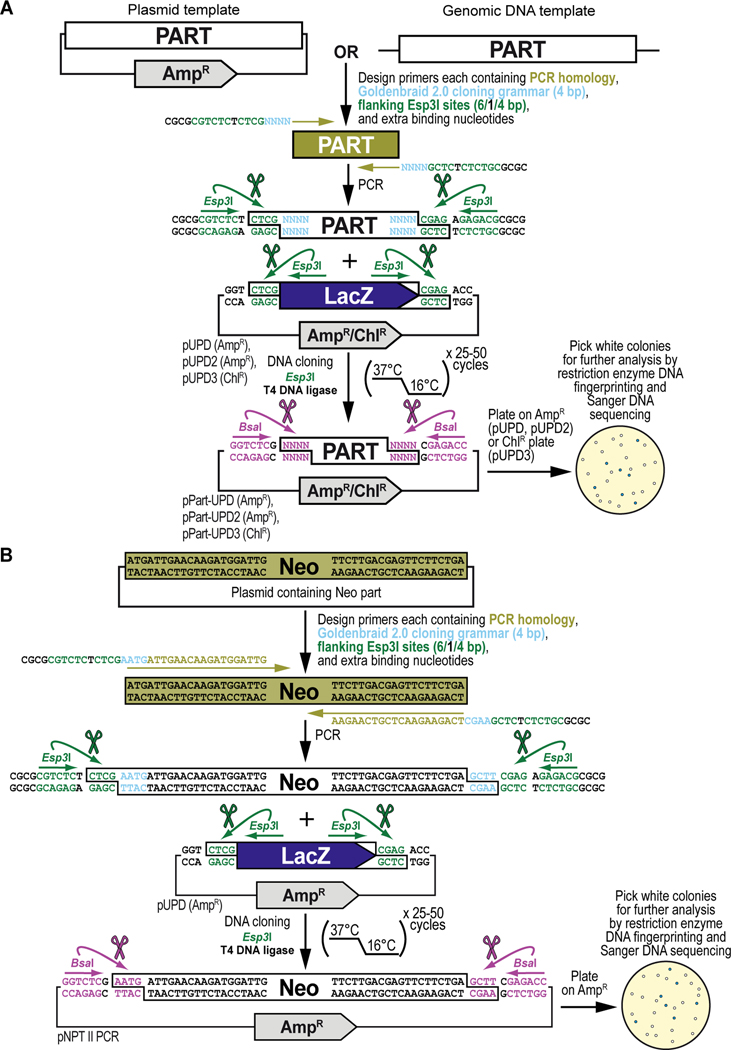
Figure 6. Obtaining and cloning a DNA part by PCR from existing DNA resources for synthetic assembly cloning. (A) General principles of obtaining and cloning a DNA part by PCR into a pUPD vector backbone.
DNA sequences from existing plasmids or genomic DNA can be integrated into the GoldenBraid2.0 synthetic DNA workflow using PCR amplification. Primers are designed that consist of 20–25 bases homologous to the DNA sequence to be amplified (PCR homology, shown in light green), and 19-nucleotide overhangs which add the desired 4-nucleotide GB2.0 cloning grammar (“NNNN”, shown in light blue) (see Figure 2B), as well as a pair of inverted Esp3I sites (shown in dark green) that will generate “CTCG” overhangs required for domestication (see below), and 4 extra nucleotides for improved restriction enzyme binding (“CGCG”, shown in black). Once PCR-amplified, the DNA fragment is combined with a Universal Part Domesticator plasmid (pUPD, pUPD2 or pUPD3), the Type IIs restriction enzyme Esp3I, and T4 DNA ligase (including 10x T4 DNA ligase buffer). The assembly protocol cycles 25 to 50 times between 37°C (favoring cutting using Esp3I) and 16°C (favoring ligation using T4 DNA ligase). After overnight selection on bacterial plates (ampicillin for pUPD and pUPD2, or chloramphenicol for pUPD3), assembled plasmids are identified as white colonies that are characterized further by restriction enzyme DNA fingerprinting and Sanger DNA sequencing, while religated domesticator plasmids are blue due to the presence of the colorimetric LacZ α-fragment. An application of the principles described here are illustrated below for the DNA sequence encoding the selectable marker Neomycin phosphotransferase II (see B). (B) Obtaining and cloning of the DNA part encoding Neomycin phosphotransferase II (Neo) into pUPD. Primers are designed, to amplify Neomycin phosphotransferase II (Neo), consisting of 20–21 bases homologous to the DNA sequence to be amplified (PCR homology, shown in light green), and 19-nucleotide overhangs which will add the desired 4-nucleotide GB2.0 cloning grammar, “AATG” to the 5’ end and “GCTT” to the 3’ end to couple it to a 5’UTR and 3’UTR, respectively (see Figure 2B), as well as a pair of inverted Esp3I sites (shown in dark green) that will generate “CTCG” overhangs required for domestication (see below), and 4 extra nucleotides for improved restriction enzyme binding (“CGCG”, shown in black). Once PCR-amplified, the “Neo” DNA fragment is combined with the Universal Part Domesticator plasmid pUPD, the Type IIs restriction enzyme Esp3I, and T4 DNA ligase (including 10x T4 DNA ligase buffer). The assembly protocol cycles 25 to 50 times between 37°C (favoring cutting using Esp3I) and 16°C (favoring ligation using T4 DNA ligase). After overnight selection on bacterial plates supplemented with ampicillin for pUPD, assembled plasmids are identified as white colonies that are characterized further by restriction enzyme DNA fingerprinting and Sanger DNA sequencing (see Text), while religated domesticator plasmids are blue due to the presence of the colorimetric LacZ α-fragment.