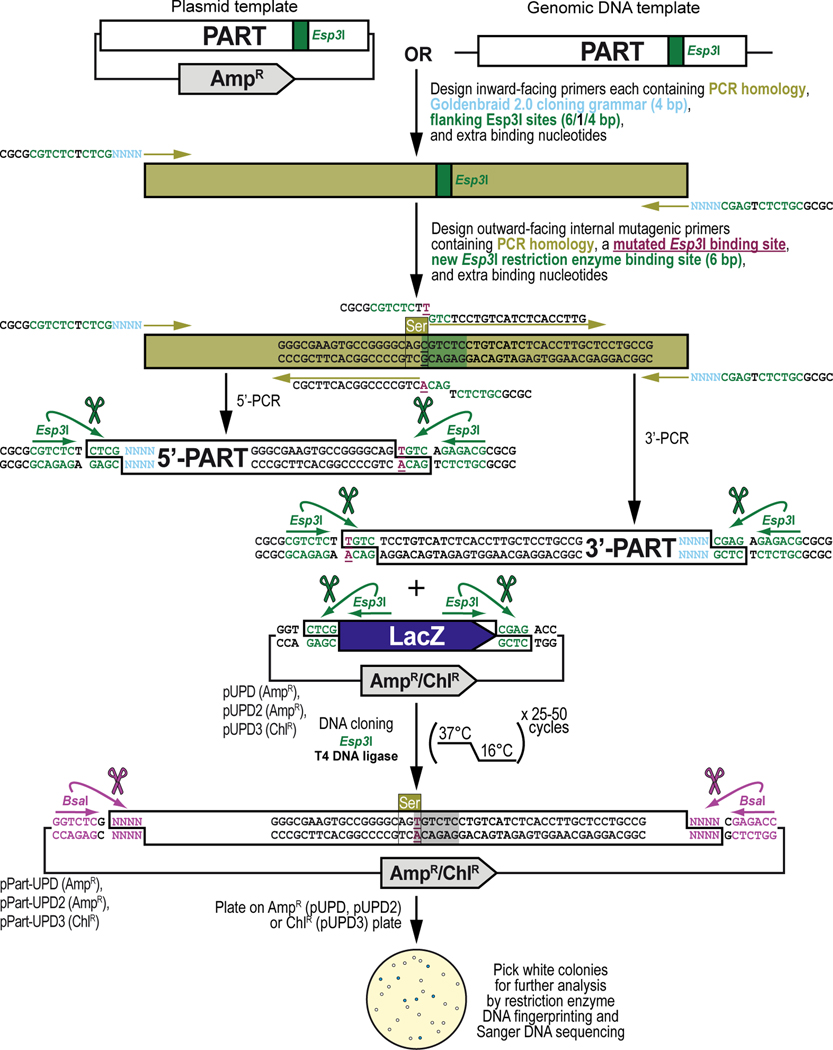
Figure 7. General principles of obtaining, adapting, and cloning a DNA part by PCR into a pUPD vector backbone.
Some DNA sequence available from existing DNA resources may contain internal binding sites for one or both Type IIs restriction enzymes (Esp3I and/or BsaI) that are used during the Goldenbraid 2.0 workflow and, while not mandatory (see Text), ideally should be removed since they can interfere with the synthetic DNA assembly process resulting in lower efficiencies or even failure of assembly reactions. These sites are removed via PCR amplification using mutagenic primers that change one (or more) of the bases within the binding site of Esp3I (shown) or BsaI (not shown). This is straightforward in a coding sequence, as sites can be removed without changing the amino acid sequence using synonymous codon switching, but care needs to be taken when sites are removed in non-coding DNA sequence ensuring not to affect any sequence critical for the function of the DNA part that needs to be integrated within the GB2.0 assembly workflow. To make the change, one additional outward-facing primer pair is designed that overlaps with the internal binding site, in addition to the first pair of inward-facing primer pair that is designed as described in Basic Protocol 2 (see Figure 6). Besides a mutation designed to remove the internal binding site for the Type IIs restriction enzyme (shown in dark purple), the outward-facing primers also contain 20–25 bases homologous to the DNA sequence to be amplified (PCR homology, shown in light green), and overhangs which add a pair of additional inverted binding sites for Esp3I (“CGTCTC”, shown in dark green), and 4 extra nucleotides for improved restriction enzyme binding (“CGCG”, shown in black). Thus, a DNA part with a single internal binding site for a Type IIs restriction enzyme will require 2 separate PCRs resulting in a 5’ and 3’ section of the DNA part which then seamlessly reconstitute the DNA part but without the binding site for the Type IIs restriction enzyme during a GB2.0 assembly reaction in a domesticator plasmid. Once PCR-amplified, both DNA fragments (encoding the 5’ and 3’ sections of the DNA part) are combined with a Universal Part Domesticator plasmid (pUPD, pUPD2 or pUPD3), the Type IIs restriction enzyme Esp3I, and T4 DNA ligase (including 10x T4 DNA ligase buffer). The assembly protocol cycles 25 to 50 times between 37°C (favoring cutting using Esp3I) and 16°C (favoring ligation using T4 DNA ligase). After overnight selection on bacterial plates (ampicillin for pUPD and pUPD2, or chloramphenicol for pUPD3), assembled plasmids are identified as white colonies that are characterized further by restriction enzyme DNA fingerprinting and Sanger DNA sequencing (see Text), while religated domesticator plasmids are blue due to the presence of the colorimetric LacZ α-fragment. An application of the principles described here is illustrated below for the DNA binding domain of the LexA repressor from Escherichia coli (LexA DBD) (see Figure 8), used as one of the parts to build the G418 sulfate-selectable LexA transactivator plasmid (see Figure 3).