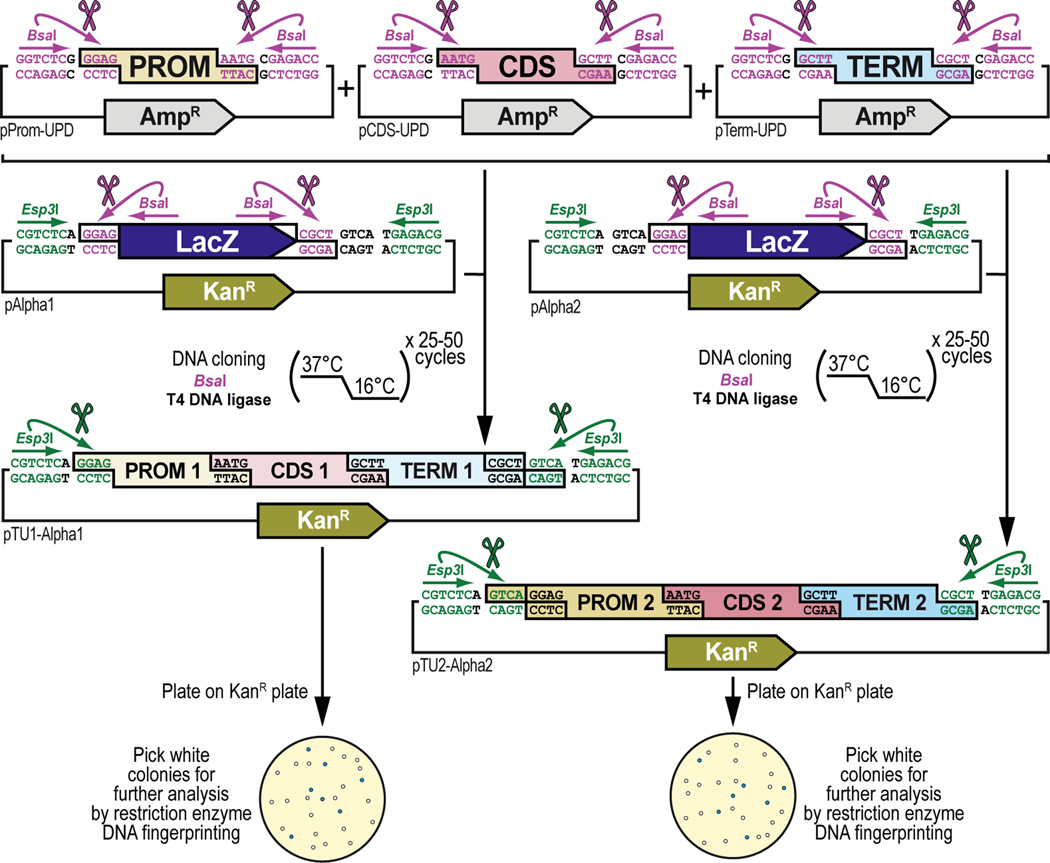
Figure 9. General principles of synthetic assembly cloning of multiple DNA parts into a multipartite transcription unit.
Transcription units are the basic functional unit of GoldenBraid2.0 synthetic DNA assembly, capable of expressing an encoded protein or with some other specific function. The simplest transcription unit (TU) consists of a promoter (PROM), coding DNA sequence (CDS), and a polyA terminator (TERM), which all exist as DNA parts cloned in a Universal Part Domesticator plasmid (pUPD, pUPD2 or pUPD3). These DNA parts are assembled in a single, one-pot cloning step using the BsaI enzyme into an Alpha destination vector. The three plasmids needed, each encoding a DNA part (pProm-UPD encoding PROM, pCDS-UPD encoding CDS, and pTerm-UPD encoding TERM), are combined with an Alpha destination vector (pAlpha1 or pAlpha2), the Type IIs restriction enzyme BsaI, and T4 DNA ligase (including 10x T4 DNA ligase buffer). The assembly protocol cycles 25 to 50 times between 37°C (favoring cutting using BsaI) and 16°C (favoring ligation using T4 DNA ligase). After overnight selection on bacterial plates (kanamycin for Alpha vectors), assembled plasmids (pTU1-Alpha1 or pTU2-Alpha2) are identified as white colonies that are characterized further by restriction enzyme DNA fingerprinting (see Text), while religated domesticator plasmids are blue due to the presence of the colorimetric LacZ α-fragment. The resulting product will consist of three DNA parts (shown), less or more (both not shown), assembled in the order dictated by their grammar identities (see Figure 2B). Different choices for promoters (PROM 1 versus PROM 2), coding DNA sequences (CDS 1 versus CDS 2), and polyA terminator (TERM 1 versus TERM 2) allows for assemblies of different transcription units (TU1 consisting of PROM 1, CDS 1 and TERM 1, versus TU2 consisting of PROM 2, CDS 2 and TERM 2) in different Alpha destination vectors (pAlpha1 versus pAlpha2, respectively) that can be brought together during further assembly steps (see Figure 11). Note the difference in the position of the internal BsaI sites in the pAlpha1 versus pAlpha2 vectors relative to the Esp3I sites. This accommodates using the same entry grammar from pUPD or Omega level vectors, while creating different exit grammars that are compatible for Omega level entry (see Figure 2C). Application of the principles described here are illustrated below (see Figure 10) for the synthetic assembly cloning of two individual transcription units used to build the G418 sulfate-selectable LexA transactivator plasmid (see Figure 3): Hsp70:CP6:Neo:TKpA (see Figure 10A and Figure 10C, left) and R70B04:dSCP:LexA-DBD:Linker:VP16-AD:SV40pA (see Figure 10B and Figure 10C, right).