Figure 5: Phylogenetic analysis of the ECSA genotype of CHIKV in Brazil.
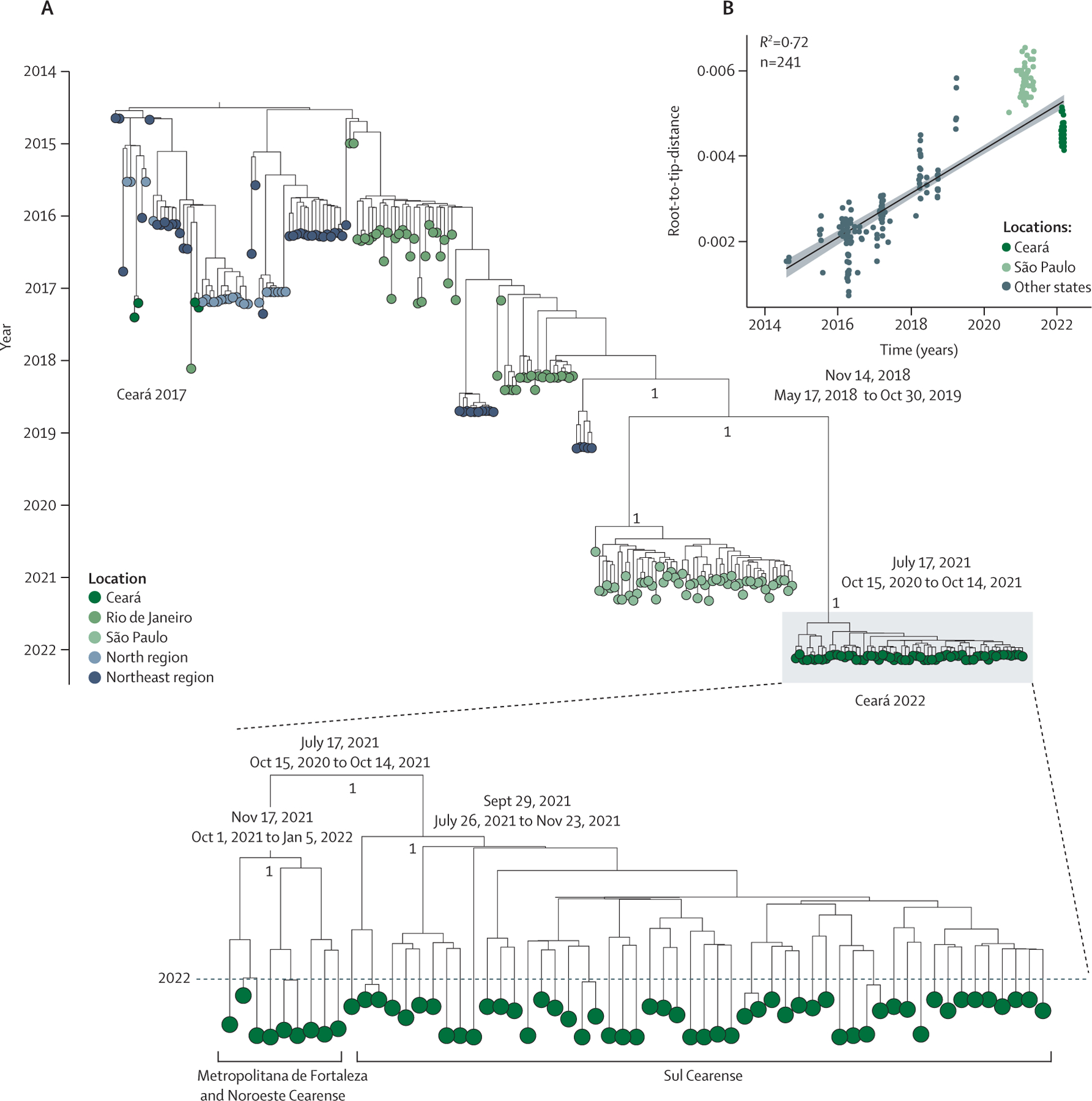
(A) Maximum clade credibility tree of 241 CHIKV genomes from the ECSA genotype, including 61 new CHIKV genomes (Ceará 2022, magnified section) from Ceará state generated in this study. Tips are coloured according to the source region or state of each sample. A strict molecular clock approach was used for generating the time-rooted tree. Posterior probability scores are shown next to key well supported nodes. Dates at key nodes are the estimated date of divergence from a common ancestor, with Bayesian credible intervals. (B) Regression of sequence sampling dates against root-to-tip genetic distances in a maximum likelihood phylogeny of the CHIKV-ECSA genotype in Brazil. Sequences are coloured according to source locations. ECSA=East-Central-South-African. CHIKV=chikungunya virus.
