ABSTRACT
Here, we report the draft genome sequence of a novel agile wallaby adenovirus that was detected in the fecal metagenome of agile wallabies. The genome is 31,512 bp long, with a G+C content of 34.4%. Currently, the pathogenic and zoonotic potential of this novel virus is unknown.
ANNOUNCEMENT
Adenoviruses are nonenveloped, icosahedral viruses with linear, unsegmented double-stranded DNA genomes (1). The genomes of adenoviruses range in size from 26 to 48 kb and commonly contain between 22 and 40 genes (1, 2). The family Adenoviridae is divided into six genera, namely, Mastadenovirus, Aviadenovirus, Atadenovirus, Siadenovirus, Ichtadenovirus, and Testadenovirus (1, 3). Adenoviral infections do not always result in disease, although they have been associated with both single and multipathogen disease processes (4–8).
The novel Agile wallaby atadenovirus 1 (AwAdV-1) described in this report belongs to the genus Atadenovirus and was originally identified in the metagenome of free-ranging agile wallabies (Notamacropus agilis) (G. R. Okoh, E. Ariel, D. Whitmore, P. F. Horwood, submitted for publication). Briefly, five fresh fecal samples were collected from the ground at grazing sites around James Cook University and Townsville University Hospital (Townsville, Australia) in 2021. The samples were homogenized, pooled, and then virally enriched by filtration (0.25 μm), ultracentrifugation (100,000 × g), and digestion with DNase I (20 U/mL). Viral DNA was then extracted using QIAamp MinElute virus kit (Qiagen). Library preparation using the Nextera DNA XT kit and Illumina sequencing (NovaSeq 6000) were performed at Macrogen (Seoul, South Korea) in paired-end 151-bp format. For this report, the sequencing reads (71,170,820) were trimmed (Trimmomatic v0.39) (9) to remove low-quality reads, normalized using BBNorm v39.01 (https://sourceforge.net/projects/bbmap/), and de novo assembled using SPAdes v3.15.5 in “careful” mode (10). The resulting contigs were searched using Diamond BLASTX (11) against the NCBI nonredundant (nr) protein database to identify the contigs corresponding to adenovirus. To assemble the genome, the reads were mapped to the identified adenoviral contig using Geneious v11.1.5 (https://www.geneious.com). Prediction of open reading frames (ORFs) was performed using Glimmer3 in Geneious v11.1.5, and ORF annotations were determined by conducting a BLASTX search against the NCBI nr protein databases (12). All tools were run with default parameters unless otherwise specified.
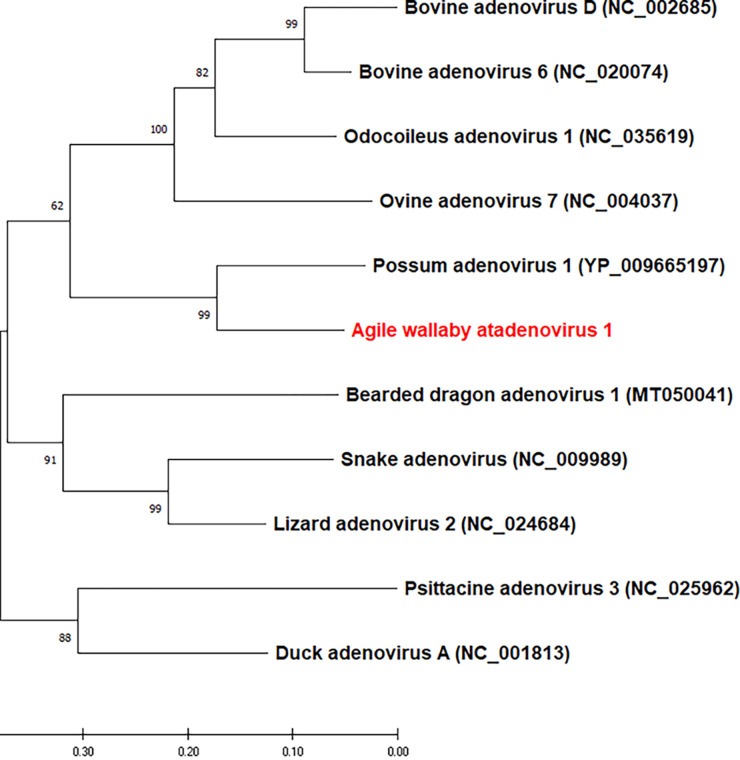
The assembled genome of AwAdV-1 was found to be 31,512 bp long, with a coverage depth of 22× and 34.4% G+C content. The genome was predicted to contain 32 ORFs with an orientation typical of atadenoviruses. Of the 32 ORFs, 26 were annotated as having various similarities to the coding genes of other atadenoviruses (Tables 1). The IVa2, penton base protein, PX, and hexon genes showed the highest amino acid identity (71% to 87%) to the reference mammalian atadenoviruses (Table 1). The AwAdV-1 genome possesses multiple insertions and deletions in most of the genes except IVa2, pX, pVI, pVIII, and U-exon. Two fiber genes, namely, fiber and IV-1 (homologous to the fiber 2 gene in lizard adenovirus 2), were present in the genome of AWAdV-1, instead of the single long fiber gene in mammalian atadenoviruses. Phylogenetic analysis based on the full amino acid sequence of penton base protein showed that AwAdV-1 belongs to the genus Atadenovirus and forms a distinct cluster with another marsupial adenovirus known as possum adenovirus 1 (Fig. 1).
TABLE 1.
Sequence comparison with the genomes of mammalian atadenoviruses
| ORF | Description (putative) | Data for virus (GenBank accession no.):a |
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Agile wallaby atadenovirus 1 |
Ovine adenovirus (NC_004037) |
Bovine adenovirus D (NC_002685) |
Bovine adenovirus E (NC_020074) |
Odocoileus adenovirus 1 (NC_035619) |
|||||||||||
| Length (nt) | Length (aa) | Length (nt) | Length (aa) | Amino acid identity (%) | Length (nt) | Length (aa) | Amino acid identity (%) | Length (nt) | Length (aa) | Amino acid identity (%) | Length (nt) | Length (aa) | Amino acid identity (%) | ||
| orf0001 | Hypothetical protein | 255 | 85 | No data | No data | No data | No data | No data | No data | No data | No data | No data | No data | No data | No data |
| orf0002 | Hypothetical protein | 402 | 134 | No data | No data | No data | No data | No data | No data | No data | No data | No data | No data | No data | No data |
| orf00003 | p32K | 1,032 | 344 | 861 | 286 | 39 | 819 | 272 | 45 | 891 | 296 | 40 | 930 | 309 | 35 |
| orf00004 | LH1 | 396 | 132 | 363 | 120 | 29 | 378 | 125 | 29 | 378 | 125 | 26 | 393 | 130 | 25 |
| orf00005 | E1B 55K | 1,293 | 431 | 1149 | 382 | 43 | 1,161 | 386 | 42 | 1,152 | 383 | 45 | 1,146 | 381 | 43 |
| orf00006 | IVa2 | 903 | 301 | 984 | 327 | 75 | 966 | 321 | 74 | 966 | 321 | 72 | 1,209 | 402 | 72 |
| orf00007 | Pol | 3,243 | 1,081 | 3,216 | 1,071 | 58 | 3,222 | 1,073 | 58 | 3,222 | 1,073 | 57 | 3,228 | 1,075 | 58 |
| orf00008 | pTP | 1,773 | 591 | 1,788 | 595 | 50 | 1,803 | 600 | 51 | 1,803 | 600 | 51 | 1,800 | 599 | 51 |
| orf00009 | 52K | 987 | 329 | 1,008 | 335 | 62 | 1,059 | 343 | 62 | 1,035 | 344 | 60 | 1,014 | 337 | 58 |
| orf00010 | pIIIa | 1,767 | 589 | 1,707 | 568 | 53 | 1,722 | 573 | 54 | 1,551 | 516 | 54 | 1,749 | 582 | 53 |
| orf00011 | Penton base protein | 1,347 | 449 | 1,359 | 452 | 68 | 1,353 | 450 | 67 | 1,359 | 452 | 68 | 1,353 | 450 | 71 |
| orf00012 | pVII | 345 | 115 | 336 | 111 | 54 | 360 | 119 | 57 | 357 | 118 | 56 | 354 | 117 | 51 |
| orf00013 | pX | 102 | 34 | 216 | 71 | 87 | 216 | 71 | 81 | 219 | 72 | 0 | No data | No data | No data |
| orf00014 | pVI | 669 | 223 | 666 | 221 | 54 | 603 | 200 | 57 | 612 | 203 | 56 | 678 | 225 | 54 |
| orf00015 | Hexon | 2,730 | 910 | 2,736 | 911 | 74 | 2,733 | 910 | 72 | 2,733 | 910 | 76 | 2,733 | 910 | 74 |
| orf00016 | 23K endoprotease | 606 | 202 | 606 | 201 | 59 | 606 | 201 | 60 | 606 | 201 | 61 | 606 | 201 | 60 |
| orf00018 | DNA binding protein | 999 | 333 | 1,149 | 382 | 56 | 1,143 | 380 | 58 | 1,140 | 379 | 56 | 1,158 | 385 | 57 |
| orf00020 | 100K | 2,055 | 685 | 1,878 | 625 | 54 | 1,887 | 628 | 56 | 1,887 | 628 | 54 | 1,914 | 637 | 55 |
| orf00021 | 33k | 512 | 170 | 402 | 133 | 39 | 405 | 134 | 65 | 408 | 135 | 39 | 414 | 137 | 38 |
| orf00022 | pVIII | 780 | 260 | 654 | 217 | 44 | 669 | 222 | 44 | 672 | 223 | 45 | 681 | 226 | 49 |
| Orf00023 | U-exon | 165 | 55 | 177 | 58 | 35 | 165 | 54 | 54 | 165 | 54 | 54 | 165 | 54 | 54 |
| orf00024 | Fiber | 891 | 297 | 1,632 | 543 | 36 | 1,608 | 535 | 36 | 1,332 | 443 | 43 | 1,422 | 473 | 29 |
| orf00025 | IV-1 | 1,554 | 518 | No data | No data | No data | No data | No data | No data | No data | No data | No data | No data | No data | No data |
| orf00026 | E4.3 | 600 | 200 | 714 | 237 | 34 | 654 | 217 | 38 | 657 | 218 | 38 | 705 | 234 | 33 |
| orf00027 | E4.2 | 678 | 226 | 663 | 220 | 40 | 678 | 219 | 42 | 660 | 219 | 42 | 678 | 219 | 39 |
| orf00028 | E4.1 | 441 | 147 | 429 | 142 | 36 | 429 | 142 | 37 | 429 | 142 | 38 | 429 | 142 | 30 |
| orf00029 | RH0 | 381 | 127 | No data | No data | No data | 564 | 187 | 45 | No data | No data | No data | No data | No data | No data |
| orf00030 | RH5 | 606 | 202 | 597 | 198 | 26 | 624 | 207 | 27 | 651 | 216 | 24 | 624 | 207 | 34 |
| orf00031 | Hypothetical protein | 372 | 124 | No data | No data | No data | No data | No data | No data | No data | No data | No data | No data | No data | No data |
| orf00032 | Hypothetical protein | 201 | 67 | No data | No data | No data | No data | No data | No data | No data | No data | No data | No data | No data | No data |
| orf00033 | Hypothetical protein | 366 | 122 | No data | No data | No data | No data | No data | No data | No data | No data | No data | No data | No data | No data |
| orf00034 | Hypothetical protein | 372 | 124 | No data | No data | No data | No data | No data | No data | No data | No data | No data | No data | No data | No data |
aa, amino acids.
FIG 1.
Phylogenetic analysis of Agile wallaby atadenovirus 1 (shown in red), based on the amino acid sequence of penton-based protein. All the sequences used in this analysis belong to the genus Atadenovirus. Following multiple sequence alignment of amino acid sequences using the Muscle program in Geneious v11.1.5, a maximum likelihood phylogenetic tree was constructed using MEGA X (13) with the best model of amino acid substitution (LG + G) and 1,000 bootstrap replications.
The pathogenic potential of AwAdV-1 is unclear; however, it could be a suitable candidate for future research in vaccinology, diagnostics, and therapeutics.
Data availability.
The raw sequence reads for this study have been deposited in the NCBI SRA database under BioProject accession number PRJNA907146 and BioSample accession number SAMN31952915. The novel genome sequence has been deposited at GenBank under the accession number OQ792214.
ACKNOWLEDGMENTS
We thank the Australian Wildlife Society and College of Public Health, Medical and Veterinary Sciences, James Cook University, for funding this research.
Contributor Information
God’spower Richard Okoh, Email: godspower.okoh@my.jcu.edu.au.
Paul F. Horwood, Email: paul.horwood@jcu.edu.au.
Jelle Matthijnssens, Katholieke Universiteit Leuven.
REFERENCES
- 1.Madarame H, Uchiyama J, Tamukai K, Katayama Y, Osawa N, Suzuki K, Mizutani T, Ochiai H. 2019. Complete genome sequence of an adenovirus-1 isolate from an African pygmy hedgehog (Atelerix albiventris) exhibiting respiratory symptoms in Japan. Microbiol Resour Announc 8:e00695-19. doi: 10.1128/MRA.00695-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Davison AJ, Benkő M, Harrach B. 2003. Genetic content and evolution of adenoviruses. J Gen Virol 84:2895–2908. doi: 10.1099/vir.0.19497-0. [DOI] [PubMed] [Google Scholar]
- 3.Böszörményi KP, Podgorski II, Vidovszky MZ, Sós E, Benkő M, Harrach B. 2020. Full genome sequence analysis of a novel adenovirus from a captive polar bear (Ursus maritimus). Virus Res 277:197846. doi: 10.1016/j.virusres.2019.197846. [DOI] [PubMed] [Google Scholar]
- 4.Jesse ST, Ciurkiewicz M, Siesenop U, Spitzbarth I, Osterhaus ADME, Baumgärtner W, Ludlow M. 2022. Molecular characterization of a bovine adenovirus type 7 (bovine atadenovirus F) strain isolated from a systemically infected calf in Germany. Virol J 19:89. doi: 10.1186/s12985-022-01817-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fent GM, Fulton RW, Saliki JT, Caseltine SL, Lehmkuhl HD, Confer AW, Purdy CW, Briggs RE, Loan RW, Duff GC. 2002. Bovine adenovirus serotype 7 infections in postweaning calves. Am J Vet Res 63:976–978. doi: 10.2460/ajvr.2002.63.976. [DOI] [PubMed] [Google Scholar]
- 6.Thomson D, Meers J, Harrach B. 2002. Molecular confirmation of an adenovirus in brushtail possums (Trichosurus vulpecula). Virus Res 83:189–195. doi: 10.1016/s0168-1702(01)00437-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Marschang RE. 2019. Emerging reptile viruses, p 267–273. In Miller RE, Lamberski N, Calle PP (ed), Fowler's zoo and wild animal medicine: current therapy, vol 9. Elsevier, St. Louis, MO. [Google Scholar]
- 8.Chen EC, Yagi S, Kelly KR, Mendoza SP, Tarara RP, Canfield DR, Maninger N, Rosenthal A, Spinner A, Bales KL, Schnurr DP, Lerche NW, Chiu CY. 2011. Cross-species transmission of a novel adenovirus associated with a fulminant pneumonia outbreak in a new world monkey colony. PLoS Pathog 7:e1002155. doi: 10.1371/journal.ppat.1002155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Buchfink B, Reuter K, Drost HG. 2021. Sensitive protein alignments at tree-of-life scale using DIAMOND. Nat Methods 18:366–368. doi: 10.1038/s41592-021-01101-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sayers EW, Beck J, Bolton EE, Bourexis D, Brister JR, Canese K, Comeau DC, Funk K, Kim S, Klimke W, Marchler-Bauer A, Landrum M, Lathrop S, Lu Z, Madden TL, O’Leary N, Phan L, Rangwala SH, Schneider VA, Skripchenko Y, Wang J, Ye J, Trawick BW, Pruitt KD, Sherry ST. 2021. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res 49:D10–D17. doi: 10.1093/nar/gkaa892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: Molecular Evolutionary Genetics Analysis across computing platforms. Mol Biol Evol 35:1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The raw sequence reads for this study have been deposited in the NCBI SRA database under BioProject accession number PRJNA907146 and BioSample accession number SAMN31952915. The novel genome sequence has been deposited at GenBank under the accession number OQ792214.