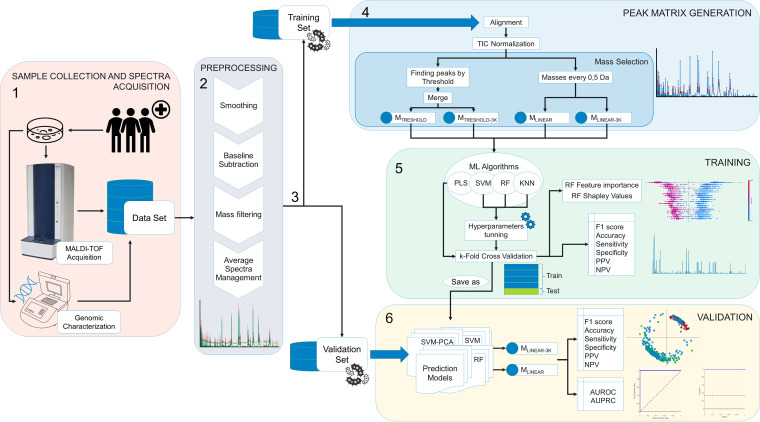
FIG 1.
Workflow of MALDI-TOF MS-based CPK prediction. 1. Sample collection and spectra acquisition. Samples are taken from infected patients and pathogens are cultured and further genotypically characterized. The same isolates are analyzed via MALDI-TOF MS. 2. Preprocessing. The mass spectra profiles are extracted from the MALDI-TOF MS and preprocessed using the Clover MS data analysis software package by applying smoothing, baseline subtraction, mass filtering, and average spectra management. 3. Data splitting. The data are split into training and validation sets. The data are labeled as CPK or NCPK as well as by the type of carbapenemase. Information regarding the sequence type, sample type, and location of origin is defined. 4. Peak matrix generation. Spectra are aligned and normalized via the TIC method. Two different mass selection methods are applied (MTHRESHOLD and MLINEAR) in two different mass ranges: one from 2,000 to 20,000 m/z and the other from 3,000 to 20,000 m/z. 5. Training. The peak matrices were used as the input data to four supervised machine learning algorithms: partial least squares discriminant analysis (PLSDA), support vector machine (SVM) with and without a principal components analysis (PCA), k-nearest neighbor (KNN) with and without a neighbourhood components analysis (NCA), and random forest (RF). The algorithms were trained and the hyperparameters were optimized. The training steps were then evaluated by calculating the resulting metrics from a k-fold cross-validation method. Once the algorithms were trained and evaluated, a prediction model for each method and peak matrix was built. The metrics reported are the accuracy, the F1 score, the sensitivity, the specificity, and the positive and negative prediction values. The contributions of individual features to the CPK prediction were determined using feature importance and Shapley values. 6. Validation. The evaluation metrics are the same as those used in the training set, but predictive performance is also measured using AUROC and AUPRC. The use of MALDI-TOF MS coupled with machine learning algorithmic processing enables the identification of CPK with an accuracy of 97.83% and of the type of carbapenemase with an accuracy of 95.24%.