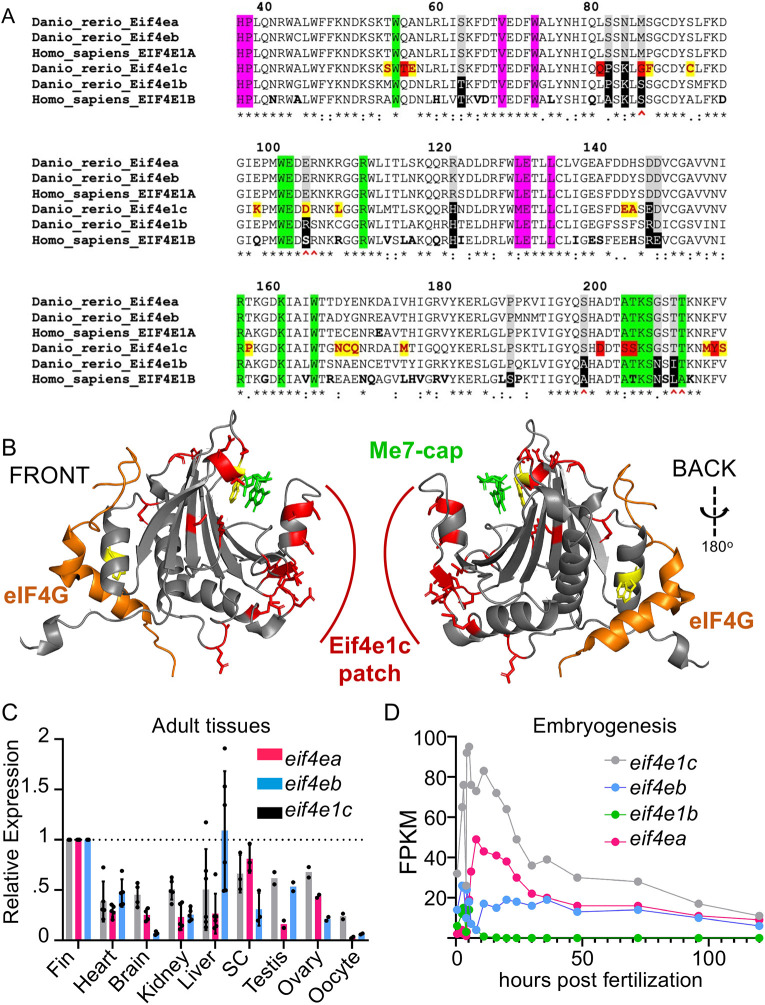
Fig. 2.
Highly conserved Eif4e1c-specific amino acids form a novel patch along the surface of the protein. (A) ClustalW sequence alignment of human versus fish Eif4E orthologs. Highlighted are universally conserved amino acids required for cap binding (green) and eIF4G recruitment (pink). Residues that distinguish EIF4E1A family members (gray) from EIF4E1B family members (black) are also highlighted. The six amino acids most responsible for strong cap binding are marked by red carets. The Eif4e1c family conserved residues are highlighted yellow with red letters. The seven residues that are identical in all EIF4E1A and EIF4E1B family orthologs throughout evolution but are uniquely changed in Eif4e1c family members are highlighted in red. Amino acids that diverge between mouse and human are bolded in the human sequence. (B) PyMOL rendition of zebrafish Eif4e1c mapped along the human structure of EIF4E1A bound to a methylated cap (green) and an EIF4G peptide (orange). The tryptophans required for both associations are conserved and highlighted in yellow. All Eif4e1c-specific residues are highlighted in red. (C) The abundance of each of the transcripts shown in the key were derived from RNAseq datasets from different adult tissues. Expression is determined relative to mob4. The FPKM for the three orthologs were similar in the fin. Values shown have been normalized to expression levels in the fin. Where error bars are presented, at least three biological replicates were used to determine the average and s.d. Only testis, ovary and oocyte had two replicates. Dots show individual data points. SC, spinal cord. (D) Time course of RNAseq of different developmental stages in embryos. Normalized sequencing reads for the four different EIF4E1 orthologs are shown.