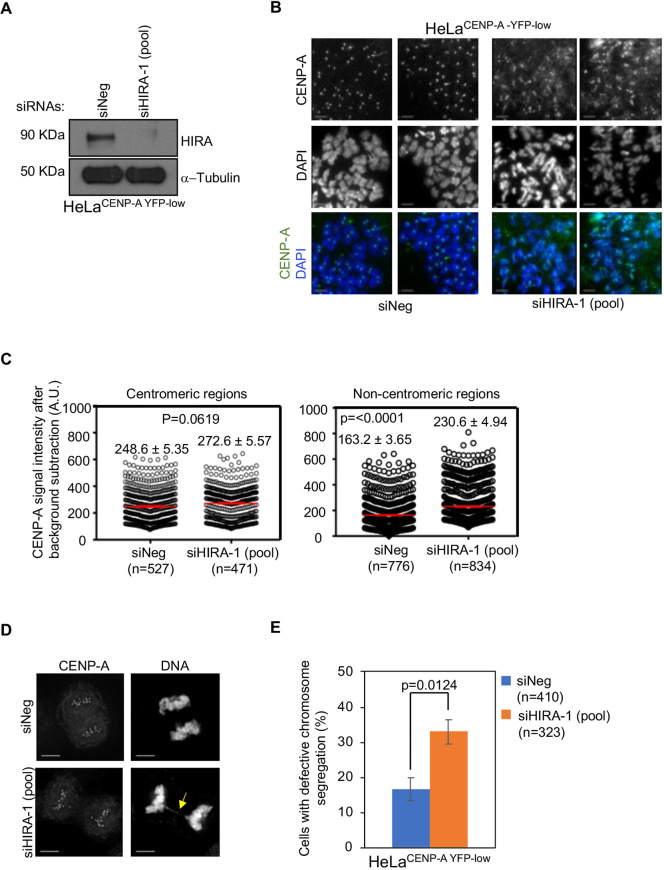
Fig. 1.
Depletion of HIRA, a positive control for the imaging-based screen, shows the mislocalization of CENP-A and CIN phenotypes in HeLaYFP–CENP-A-low cells. (A) Western blots of lysates prepared from HeLaYFP–CENP-A-low cells transfected with the indicated siRNAs for 72 h and analyzed using the antibodies as indicated. (B) Representative images of metaphase chromosome spreads showing the localization of CENP-A at centromeric and non-centromeric regions in HeLaYFP–CENP-A-low cells transfected with the indicated siRNAs. Metaphase chromosome spreads were prepared 72 h post transfection, and cells were immunostained using an antibody against CENP-A and stained with DAPI. Scale bars: 5 µm. (C) Quantification of CENP-A signal intensities at centromeric (left) and non-centromeric (right) regions in metaphase chromosome spreads of HeLaYFP–CENP-A-low cells transfected with the indicated siRNAs. Each circle represents a spot on a centromeric or non-centromeric region. n denotes the number of chromosomes analyzed. Red lines indicated mean±s.e.m. for YFP signal intensities across areas measured in the number of spots indicated as n from three independent experiments. A.U., arbitrary units. (D,E) Immunostained images (D) and bar chart (E) showing the proportion of cells with defective chromosome segregation in HeLaYFP–CENP-A-low cells transfected with the indicated siRNAs. The yellow arrow shows lagging chromosomes. Error bars represent s.e.m. across three independent experiments. n denotes the number of cells analyzed from three experiments. P-values shown in C, E were calculated using unpaired, two-tailed t-test.