Fig. 1. Differentiating antiviral effector CD8+ T cells exhibit highly dynamic accessible chromatin and cBAF occupancy patterns.
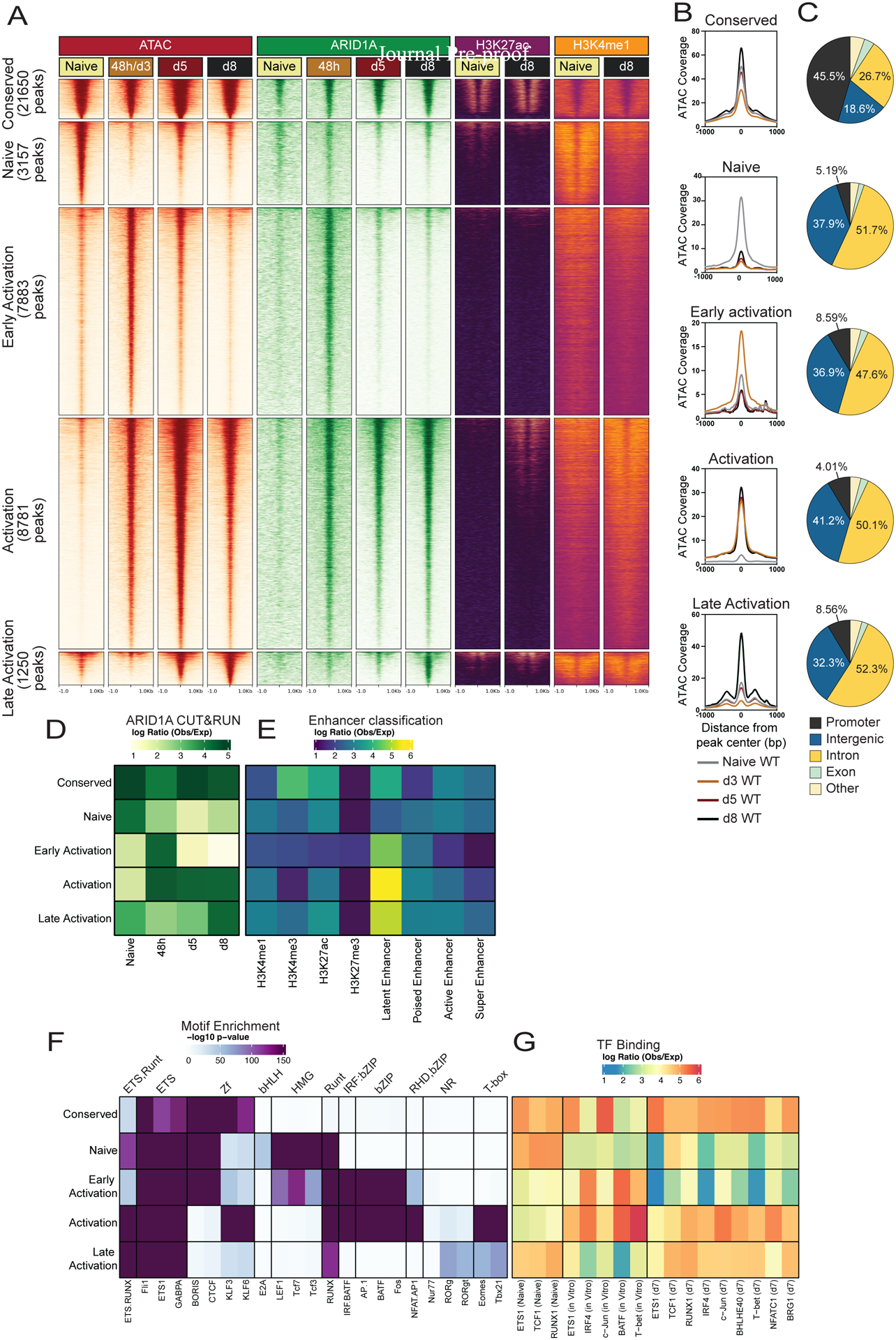
(A) Signal tracks of ATAC-seq, ARID1A C&R, H3K27ac ChIP-Seq and H3K4me1 ChIP-Seq (GSE89036) in naïve, 48h in vitro activated, or d3, d5, or d8 P14 CD8+ T cells from LCMV-Armstrong infection. (B) ATAC-seq signal coverage and (C) annotation of OCRs from clusters in A. (D-G) Using the OCRs from clusters in (A) we correlated those with ARID1A binding (D), histone modifications to define enhancers (E), predicted TF motif enrichment (F) and actual TF binding (observed/expected) of the indicated TFs from public datasets (GSE54191, GSE192390, GSE166718) (G).
