Fig 4. ARID1A-dependent OCRs in d8 MP, EEC, and TE cells are largely shared across subsets.
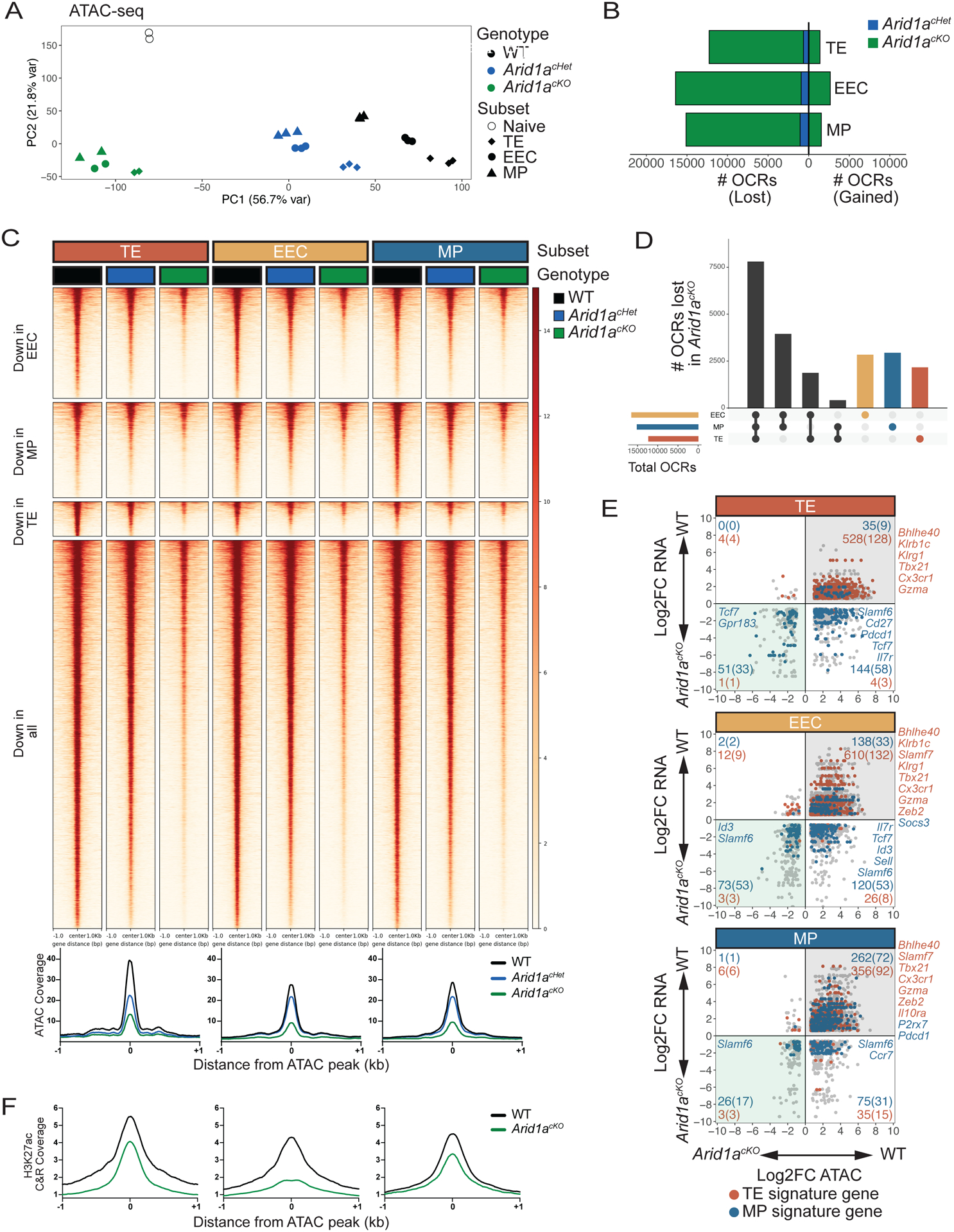
WT, Arid1acKO and Arid1acHet effector cells were isolated at d8 p.i. and sorted based on expression of TE, EEC and MP markers as defined in Figure 3A and differential OCRs were compared by ATAC-sequencing (2-fold change, adjusted p value<0.05, Benjamini-Hochberg). (A) Principal component analysis plot of ATAC-seq from WT, Arid1acHet, and Arid1acKO subsets at d8 post-infection. (B) Number of OCRs lost and gained in Arid1acKO and Arid1acHet subsets relative to WT cells. (C-D) ATAC-seq signal heatmaps of WT, Arid1acHet, and Arid1acKO d8 subsets. OCRs are clustered by whether they are lost in the Arid1acKO relative to WT cells in individual subsets or lost in all three subsets (C) and UpSet plot shows the number of shared and subset-specific OCRs lost in Arid1acKO subsets relative to WT cells (D). (E) Gene expression and paired chromatin accessibility of annotated OCRs in Arid1acKO and WT d8 subsets. TE- and MP-signature genes are highlighted in blue and red, respectively. (F) H3K27ac CUT&RUN signal coverage centered on ARID1A-dependent ATAC-seq peaks in WT vs. Arid1acKO cells TE, EEC, and MP cells.
