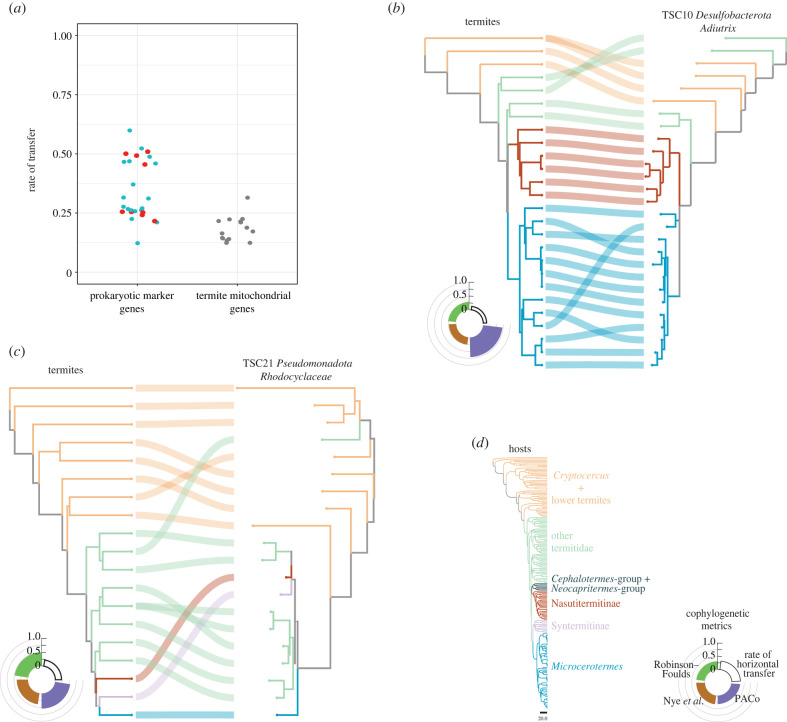
Figure 3.
Rate of transfer and phylogenetic trees of some termite-specific bacterial clades (TSCs) showing strong cophylogenetic signals with termites. The TSC phylogenetic trees were reconstructed with IQtree using the RY-recoded DNA sequence alignments from the marker gene COG0552. (a) Rates of horizontal transfer of 27 TSCs were estimated using the ML method implemented in the GeneRax software. Red dots represent bacterial clades showing no significant cophylogenetic signals, and cyan dots represent bacterial clades showing significant cophylogenetic signals. Tanglegrams between termites and (b) the Desulfobacterota Adiutrix TSC10 and (c) the Pseudomonadota Rhodocyclaceae TSC21. (d) Phylogenetic tree of termites inferred from mitochondrial genomes. The diagrams below the phylogenetic trees indicate the results of the cophylogenetic analyses and the estimation of the horizontal transfer rate. The scale bar represents substitutions per site.