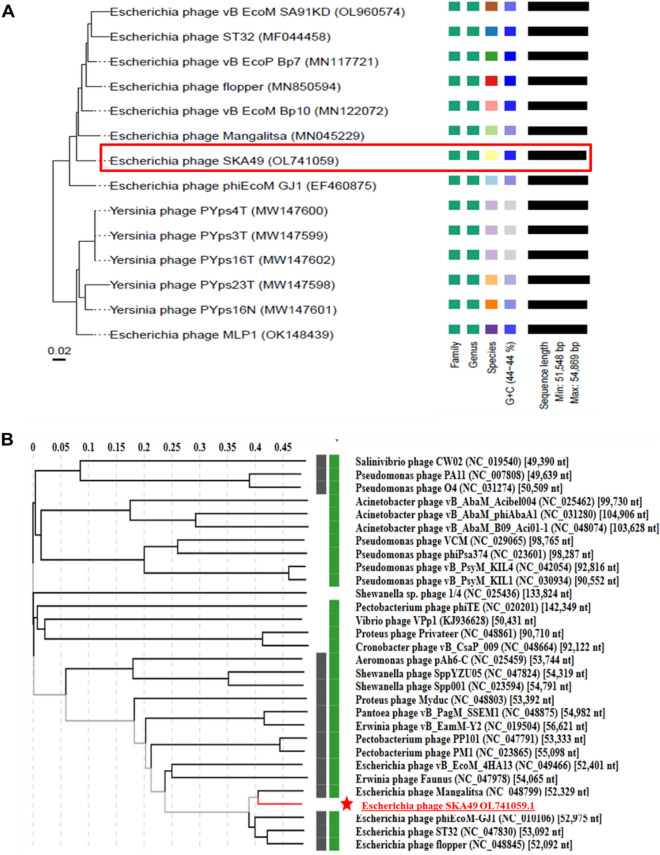
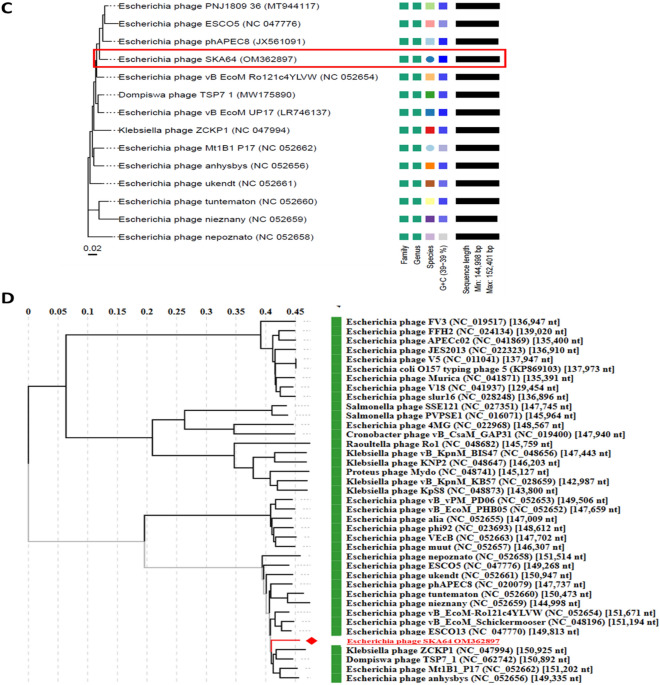
Figure 9.
(A and B) Phylogenetic position of Escherichia phage SKA49. (A) Nucleotide based GDBP tree provides an estimate of phage relationship with close homologs in BLAST n using VICTOR. Analysis identified one Family, one genus, and 12 species clusters respectively, identifying SKA49 as new species. (B) A proteome tree made by ViPTree software that uses genome wide similarities calculated by tBLASTx. Genomes for comparison were selected according to SG scores of 30 sequences in VIP tree ranging from 1 to 0.02 (supplementary Table S10B) (C and D) Phylogenetic position of Escherichia phage SKA64. (C) Nucleotide based GDBP tree provides an estimate of phage relationship with close homologs in BLAST n using VICTOR. Analysis identified one Family, one genus, and 14 species clusters respectively, identifying SKA64 as new species. (D) A proteome tree made by VIP Tree software that uses genome wide similarities calculated by tBLASTx using Similarity score generated by software . Both trees place SKA64 in the same genus. Genomes for comparison were selected according to SG scores of 39 sequences in VIP tree ranging from 1 to 0.07 (supplementary Table S10B).