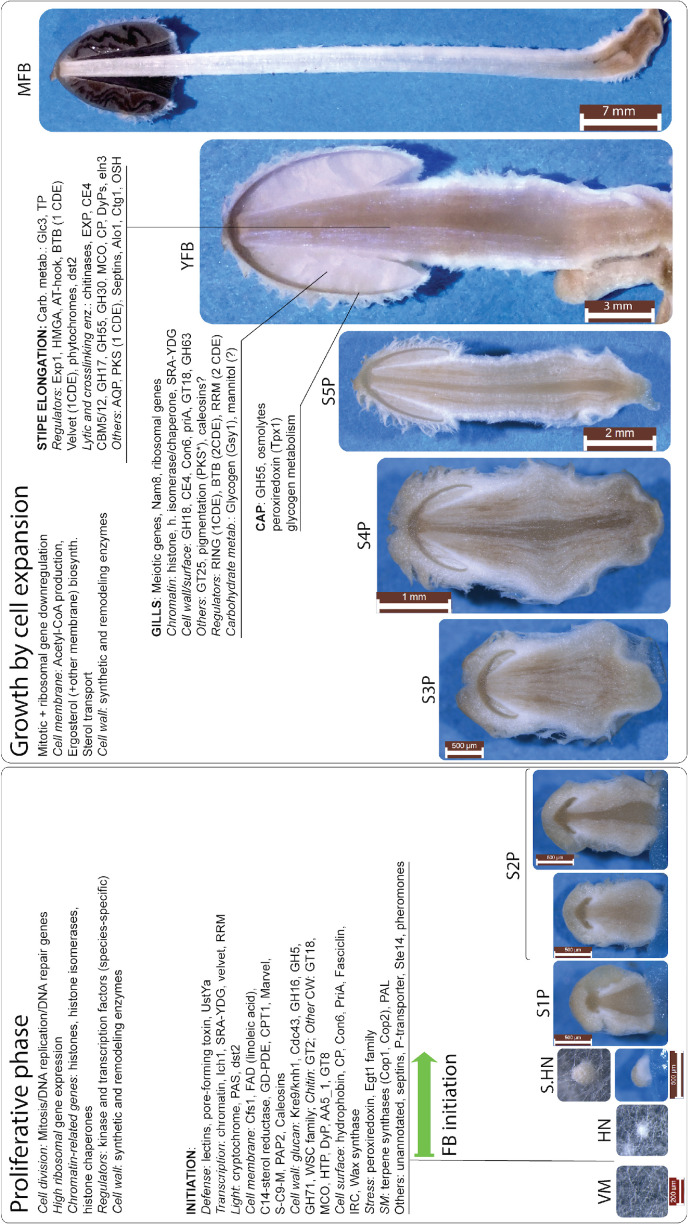
Fig. 15.
The most important, conserved genes belonging to each phase are shown - genes with limited expression conservation or species-specific expression are not depicted.
[footnote: *pigment patterns vary by species] TP: trehalose phosphorylase, PKS: pyloketide synthase; OSH: Oxysterol Binding Protein Homologue; AQP: aquaporins; EXP: expansins; MCO: multicopper oxidase; RRM: RNA recognition motif family proteins; PAP2: diacylglycerol pyrophosphate phosphatases; FAD: fatty acid desaturase; GD-PDE: Glycerophosphodiester phosphodiesterases; S-C9-M: sphingolipid C9-methyltransferases; IRC: intradiol ring cleavage dioxygenase; PAL: phenylalanine ammonia-lyase; SM: secondary metabolism; HTP: heme-thiolate peroxidase; CP: cerato-platanin. genes associated with cap autolysis are not shown on this Figure, because it is a species-specific trait of C. cinerea. Note that light induction schemes are not shown on this Figure, as this varies from species to species.