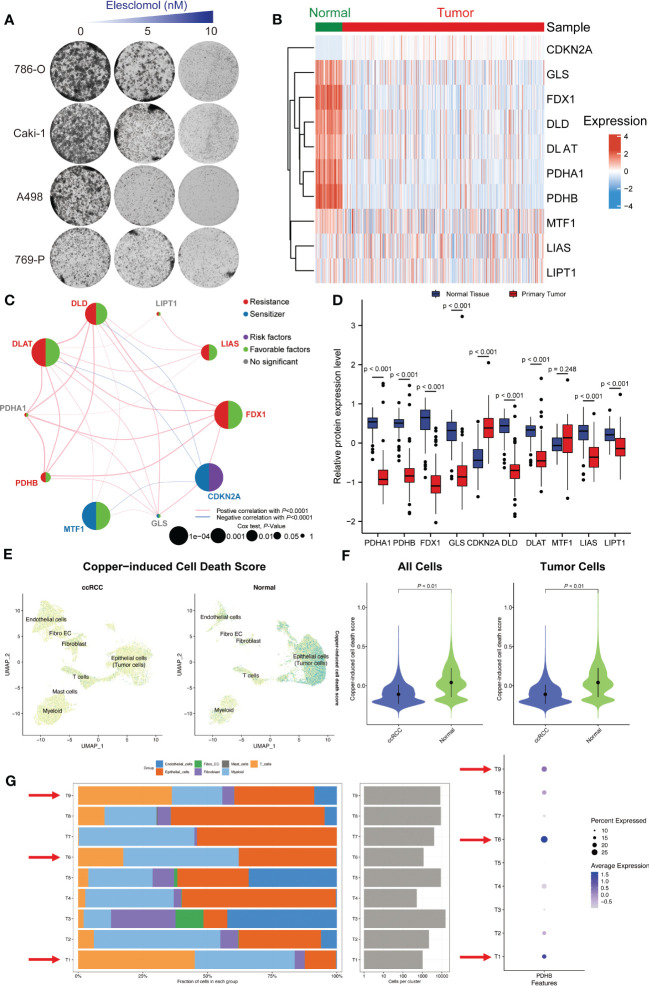
Figure 2.
Single-cell and bulk RNA-seq revealed the distinct landscape of cuproptosis regulator genes in ccRCC. (A) The cell viability of 786-O, Caki-1, A498 and 769-P cells decreased significantly after cuproptosis activator (elesclomol) treatment. (B) The expression pattern of these cuproptosis-related genes in ccRCC and adjacent normal tissues. (C) Correlation analysis of these cuproptosis-related genes. (D) The protein expression level of these cuproptosis regulators in CPTAC-ccRCC database. (E) Single-cell RNA-seq illustrated the distribution of copper-induced cell death scores by “AddModuleScore” algorithm. (F) Copper-induced cell death signature was significantly down-regulated in ccRCC tumors tissues compared with normal samples both in all cells and tumor cells. (G) The cell proportion among different patients and correlations of PDHB expression level.