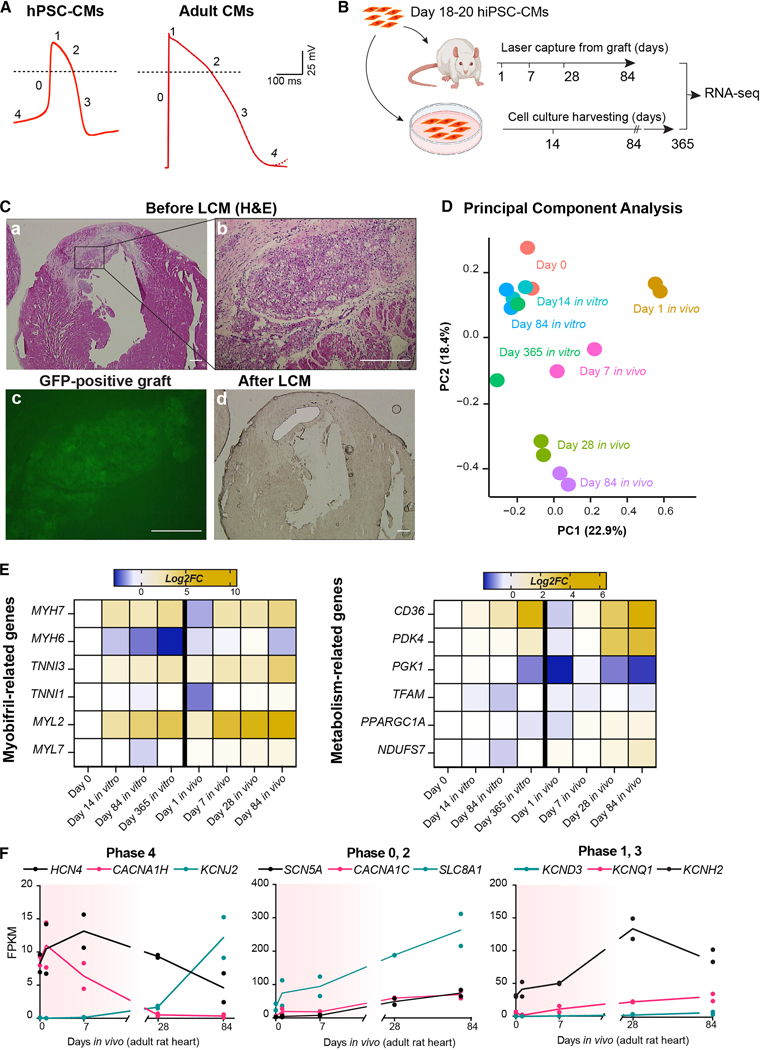
Figure 1. Gene expression analysis of hiPSC-CMs during in vivo transplantation compared to 2D culture.
(A) Representative action potential traces from hESC-CMs and adult ventricular CMs (dotted line indicates 0 mV). Adult CMs adapted from Karbassi et al.16 (B) Experimental layout for RNA-seq experiments. (C) Representative hematoxylin and eosin-stained (H&E) histological analyses of rat heart engrafted with hiPSC-CMs at day 84 after injection (a-b), and adjacent unstained sections showing the graft site before (c, fluorescence image of GCaMP3 signal) and after laser capture microdissection (LCM; d, brightfield image). Scale bar = 200 μm for a, d; 50 μm for b, c. (D) Principal component analysis (PCA) of RNA-seq data set described in B. The percentage of gene expression variance expressed by each PC is indicated. Day 0 indicates hiPSC-CMs in vitro at day 18–21 as depicted in B. (E) RNA-seq expression heatmap of selected maturation-related genes (data plotted as average log2 fold-change from day 0 for three biological replicates). (F) Selected RNA-seq data for ion channels involved in AP. Red shading indicates the approximate window of engraftment arrhythmia. See also Supplementary Figs. 1C, D.