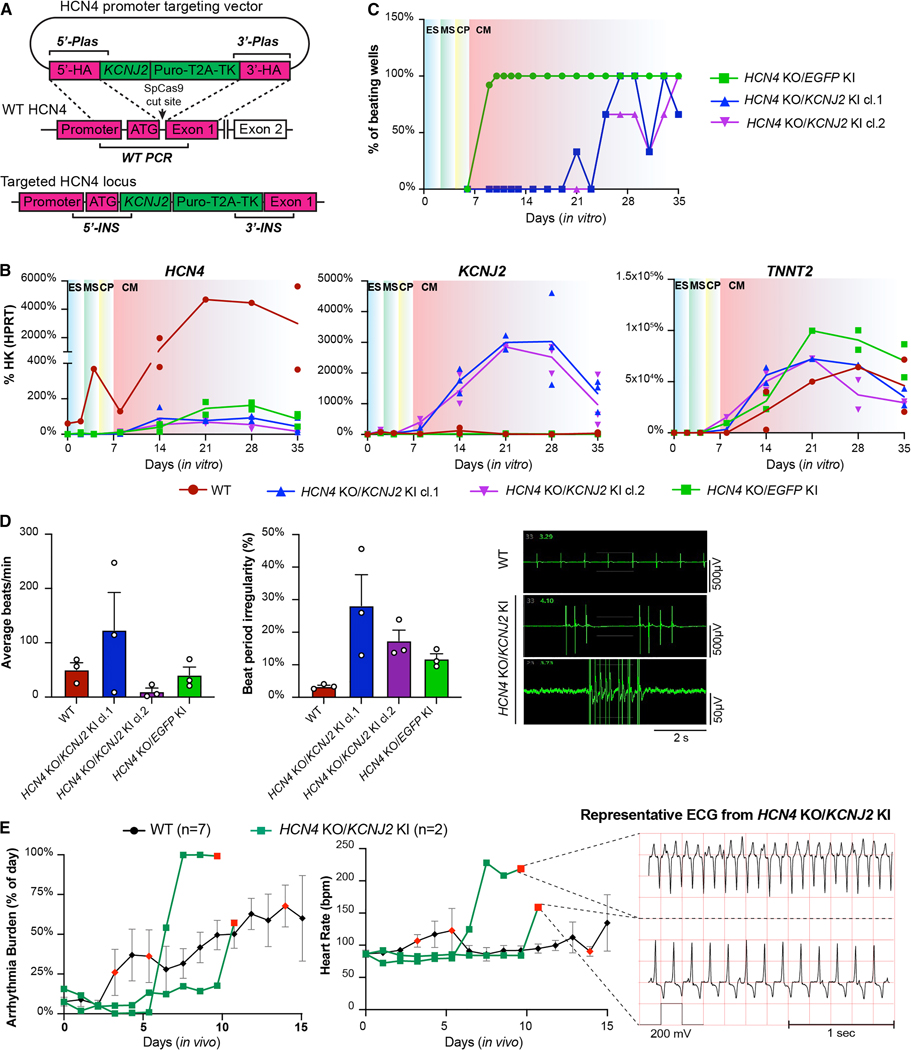
Figure 3. HCN4 and KCNJ2 perturbation is not sufficient to prevent automaticity or EA.
(A) Gene-editing approach to knock-in KCNJ2 under the transcriptional control of the HCN4 promoter in RUES2 hESCs. Genotyping PCR strategies for on- and off-target insertions are indicated; see also Supplementary Figure 4A. (B) Time course qRT-PCR analysis of HCN4, KCNJ2 and TNNT2 expression during cardiac differentiation of the indicated WT and gene-edited hESCs. ES: embryonic stem cell, MS: mesoderm, CP: cardiac progenitor, CM: cardiomyocyte. N = 2 differentiations per cell line. (C) Representative quantification of spontaneous beating during hESC-CM differentiation from HCN4 KO/KCNJ2 KI clones compared to WT. See Methods for details on quantification. (D) Spontaneous activity of HCN4 KO/KCNJ2 KI clones quantified by MEA, and representative traces. Given the marked irregularity of automaticity in these lines, data are reported as average beats in 5 min recording (left panel) and the corresponding % beat irregularity (right panel), calculated as standard deviation of the beat period record in 100 sec, divided by the mean of the beat period in that same period. Data are plotted as mean ± SEM of 3 independent experiments each with 8 technical replicates. (E) In vivo data showing EA burden (left panel) and heart rate of animals transplanted with HCN4 KO/KCNJ2 KI hESC-CMs compared to WT, with representative ECG traces on the right. Data shown for WT controls as described in Fig. 2G and individual traces (N = 2) for HCN4 KO/KCNJ2 KI. Red-colored symbol represents animals that reached prespecified EA endpoints leading to withdrawal from the study, as defined in the Methods section.