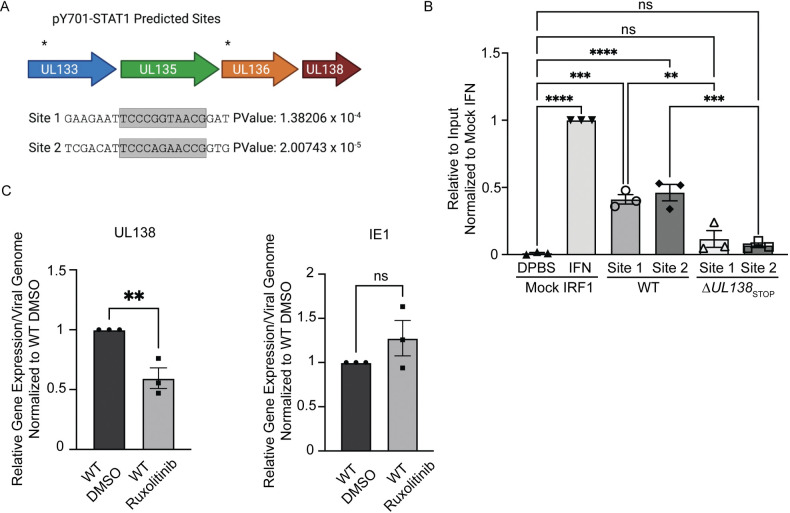
Fig 7. pSTAT1 binds to the viral genome and regulates UL138 transcription during a replicative infection.
(A) The HCMV genome was run through PhysBinder Software to obtain potential pSTAT1 consensus sites on the viral genome. Site 1 and 2 are within UL133 and UL136 respectively and are upstream of UL138. (B) Fibroblasts were infected (MOI = 1) with either a WT or ΔUL138STOP virus. At 72 hpi, uninfected cells were treated with DPBS (vehicle control) or 1000 U/mL of universal type 1 interferon for 30 minutes. After 30 minutes, all cells entered the CUT&RUN protocol developed by Cell Signaling Technologies. qPCR was performed on the input DNA and immunoprecipitated DNA utilizing primers specific to pSTAT1 binding sites of interest. Percent Immunoprecipitated was calculated to 2% of the input sample and graphed normalized to mock cells with interferon treatment. Statistical significance was calculated using One-Way Anova and represented by asterisks; **p-value <0.01, ***p-value <0.001, ****p-value <0.0001. (C) Fibroblasts were infected (MOI = 1) with a WT virus. At the time of infection, cells were treated with DMSO (vehicle control) or 5 μM Ruxolitinib every 24 hours. At 72 hpi, DNA and RNA was isolated and transcripts encoding UL138 and IE were quantified by RT-qPCR normalized to H6PD. Viral genomes were analyzed through qPCR using primers for b2.7kb and RNaseP as a loading control. Relative expression was determined using the ΔCT method and values were normalized to the DMSO control and graphed for statistical analysis. Statistical significance was calculated using unpaired student t test and represented by asterisks; **p-value <0.01. Graphs represent the mean of three replicates and error bars represent SEM.