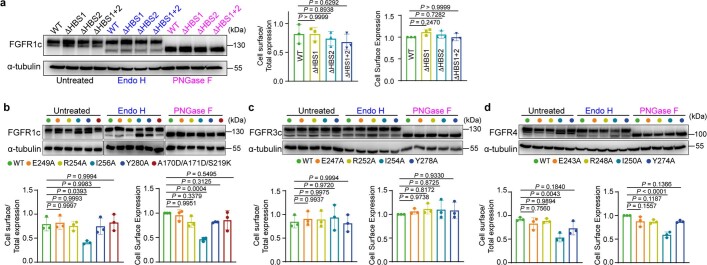
Extended Data Fig. 3. Cell surface expression analysis of mutated FGFRs via Endoglycosidase H (Endo H) sensitivity assay.
a, Denatured lysates from L6-FGFR1cWT, L6-FGFR1cK175Q/K177Q (FGFR1cΔHBS1), L6-FGFR1cK207Q/R209Q (FGFR1cΔHBS2), and L6-FGFR1cK175Q/K177Q/K207Q/R209Q (FGFR1cΔHBS1+2) cells were incubated with Endoglycosidase H (Endo H) or Peptide-N-Glycosidase F (PNGase F) or left untreated. Samples were immunoblotted with FGFR1c isoform-specific antibody. Immunoblotting data were quantitated as described in the Methods section and are presented as the mean ± SD. b–d, Denatured lysates from L6 cell lines stably expressing FGFR1cWT, its four Site 2 (FGFR1cE249A, FGFR1cR254A, FGFR1cI256A, FGFR1cY280A) and one Site 1 (FGFR1cA170D/A171D/S219D) mutants (b), wild-type FGFR3c (FGFR3cWT) or its four Site 2 mutants (FGFR3cE247A, FGFR3cR252A, FGFR3cI254A, FGFR3cY278A) (c), wild-type FGFR4 (FGFR4WT) or its four Site 2 mutants (E243A, R248A, I250A, Y274A) (d) were treated with Endo H, PNGase F or left untreated. Samples were immunoblotted with FGFR isoform-specific antibodies as indicated. Experiments were performed in biological triplicates with similar results. Quantitation was done as described in the Methods section and are presented as the mean ± S.D. P values were determined by One-way ANOVA followed by Tukey’s multiple comparisons post hoc test.