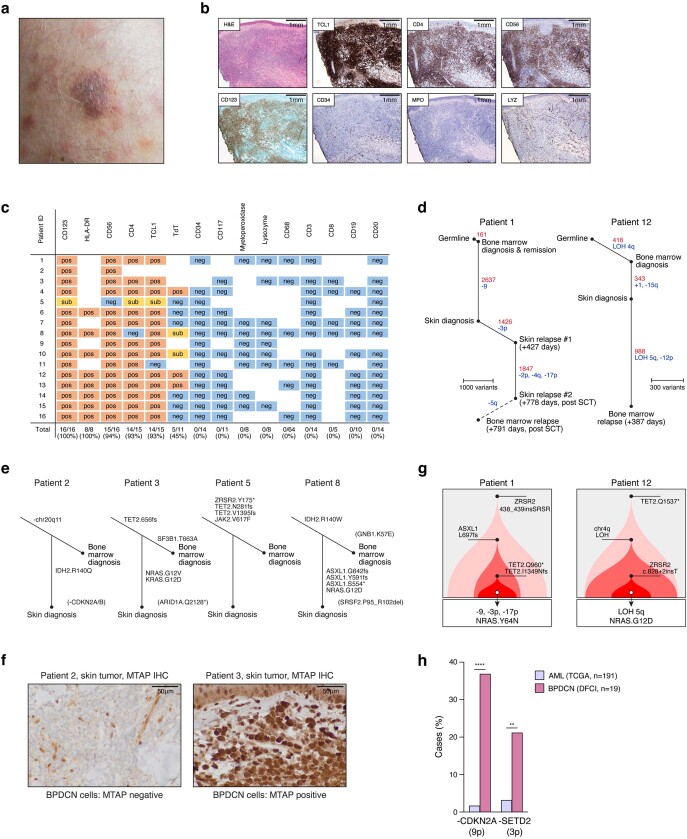
Extended Data Fig. 1. Clinical evaluation of bone marrow involvement in BPDCN.
a, Clinical photo of a representative BPDCN skin lesion at initial diagnosis showing a violaceous nodule that occurred on a background of diffuse pink, brown and violaceous papules, plaques, and tumours. b, Low-power histologic images of a representative BPDCN skin tumour showing extensive dermal infiltration by malignant BPDCN cells with characteristic immunophenotype, including positivity for TCL1, CD4, CD56, and CD123, and negativity for CD34, myeloperoxidase (MPO), and lysozyme (LYZ). These features distinguish BPDCN in the skin from histologic mimics, including acute myeloid leukaemia with monocytic differentiation and chronic myelomonocytic leukaemia. c, Immunophenotypic features of the sixteen BPDCN cases included in the cohort, as determined by immunohistochemistry and/or flow cytometry performed during initial pathology evaluation. This shows the presence of BPDCN markers (TCL1, CD4, CD56, CD123) and the absence of myeloid, T, or B-cell lineage markers that define other forms of acute leukaemia. d, Tumour phylogenies and sample-specific alterations for Patients 1 and 12 as determined by whole-genome sequencing. Somatic SNVs (red) and copy-number alterations (blue) were defined at the latest relapse time point, and then assessed in prior skin tumour and bone marrow diagnosis samples. e, Tumour phylogenies and sample-specific alterations for Patients 2, 3, 5, and 8 as determined by targeted sequencing (RHP and Oncopanel, see Methods). Mutations that are not covered in both the RHP (bone marrow samples) and Oncopanel (skin samples) assays are indicated in parentheses. f, Immunohistochemistry for MTAP protein (brown) as a marker of chr 9p21.3 loss in BPDCN skin tumours. MTAP is lost in skin tumour cells from patient 2, which contain a focal deletion on chr9p21.3, and retained in skin tumour cells from patient 3, which do not have chr9p21.3 loss. g, Inferred clonal architecture in diagnostic marrows for Patients 1 and 12 in (d). Premalignant subclones related to BPCN skin tumours are indicated with arrows, and further annotated with skin-specific progression mutations. h, Frequency of copy-loss in CDKN2A/chr9p and SETD2/chr3p in BPDCN and AML. Statistical significance by two-sided Fisher’s exact test. TCGA, The Cancer Genome Atlas. DFCI, Dana-Farber Cancer Institute. ** P < 0.01, **** P < 0.0001. Related to Fig. 1.