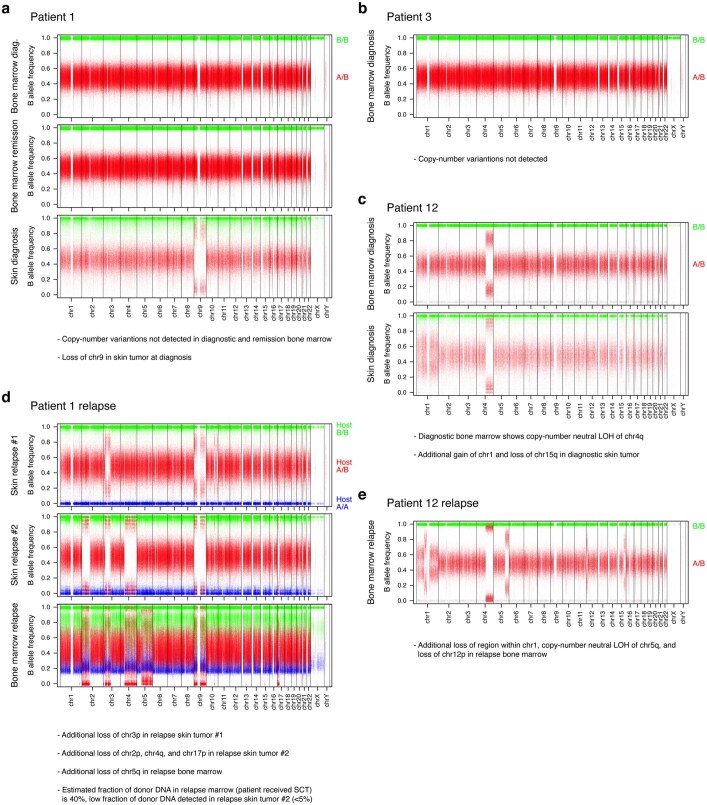
Extended Data Fig. 4. Copy-number alteration analysis of Patient 1, 3, and 12 whole-genome sequencing data.
a, Genome plots show B (minor) allele frequencies for all single-nucleotide variants (SNVs) detected in the Patient 1 germline sample using whole-genome sequencing. SNVs are shown for bone marrow samples at diagnosis and remission, and a skin tumour sample. SNVs that are heterozygous (A/B) in the germline sample are indicated in red, homozygous SNVs (B/B) are indicated in green. Loss of chr9 specific to the skin tumour sample is indicated. b, Genome plot shows similar analysis for a bone marrow sample of Patient 3, not harbouring any copy-number alterations. c, Genome plots show similar analysis for a bone marrow and skin tumour sample of Patient 12. Copy-number neutral loss of heterozygosity (LOH) on chr4q was detected in both samples. Additional loss of chr15q and gain of chr1 was detected in the skin tumour sample. d, Genome plots show single-nucleotide variants (SNVs) detected in the relapse bone marrow sample of Patient 1 collected after receiving an allogeneic stem cell transplant. SNVs are shown for skin relapse sample #1 and #2, as well as a bone marrow relapse. Samples were collected at consecutive time points. SNVs homozygous in the host germline sample are indicated in blue (A/A) and green (B/B), heterozygous SNVs are indicated in red (A/B). In addition to the loss of chr9 in the skin sample at diagnosis, additional copy-number alterations are detected in each sample: Loss of chr3p is first detected in skin relapse #1, loss of chr2p, chr4q, and chr17p are first detected in skin relapse #2, and loss of chr5q is first detected in the bone marrow relapse. Donor DNA is detected in both skin sample #2 and bone marrow relapse sample. Estimated fraction of donor DNA is <5% and 40%, respectively. e, Genome plot shows similar analysis for a bone marrow relapse sample of Patient 12, who did not receive an allogeneic stem-cell transplant. Additional alterations in chr1, chr5q, and chr12p are indicated. Related to Fig. 1.