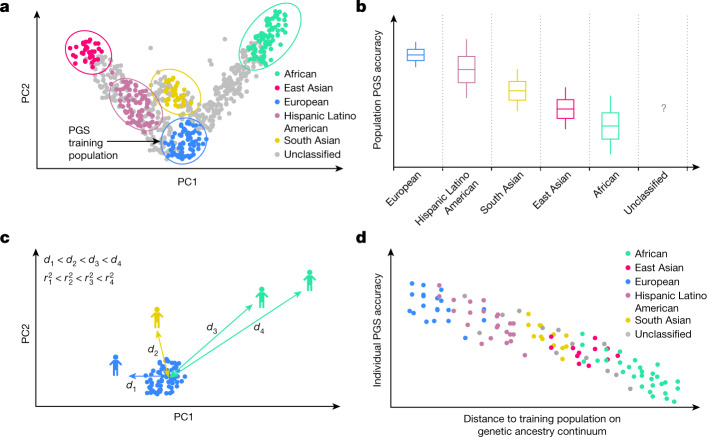
Fig. 1. Illustration of population-level versus individual-level PGS accuracy.
a, Discrete labelling of GIA with PCA-based clustering. Each dot represents an individual. The circles represent arbitrary boundaries imposed on the genetic ancestry continuum to divide individuals into different GIA clusters. The colour represents the GIA cluster label. The grey dots are individuals who are left unclassified. b, Schematic illustrating the variation of population-level PGS accuracy across clusters. The box plot represents the PGS accuracy (for example, R2) measured at the population level. The question mark emphasizes that the PGS accuracy for unclassified individuals is unknown owing to the lack of a reference group. Grey dashed lines emphasize the categorical nature of GIA clustering. c, Continuous labelling of everyone’s unique position on the genetic ancestry continuum with a PCA-based GD. The GD is defined as the Euclidean distance of an individual’s genotype from the centre of the training data when projected on the PC space of training genotype data. Everyone has their own unique GD, , and individual PGS accuracy, . d, Individual-level PGS accuracy decays along the genetic ancestry continuum. Each dot represents an individual and its colour represents the assigned GIA label. Individuals labelled with the same ancestry spread out on the genetic ancestry continuum, and there are no clear boundaries between GIA clusters. This figure is illustrative and does not involve any real or simulated data.