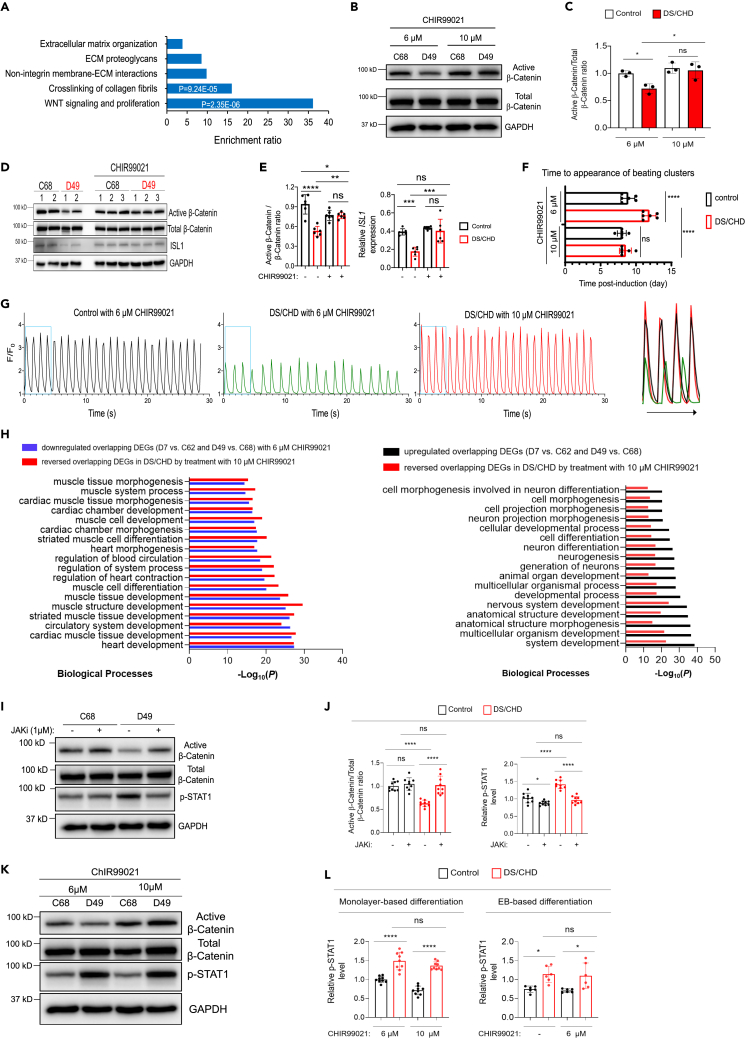
Figure 3.
Restoration of the Wnt/β-Catenin pathway normalizes cardiac differentiation of DS/CHD iPSCs
(A) REACTOME analysis of downregulated genes in DS/CHD cells on differentiation day 3. On day 3 post-induction of differentiation, RNA-seq was performed on C62, C68, D7, and D49 iPSC lines. Genes with expression ≥25 FPKM that were downregulated ≥1.5-fold (p < 0.05) were selected for REACTOME analysis.
(B) Representative immunoblotting for indicated proteins in control and DS/CHD cells on differentiation day 3. Cells were cultured with either 6 or 10 μM CHIR99021, an activator of the Wnt/β-Catenin pathway between day 0 and day 1.
(C) Quantification of the active β-Catenin/total β-Catenin ratio by immunoblotting. Each filled circle represents an individual iPSC line of 3 control (C42, C62, and C68) and 3 DS/CHD (D19, D7, and D49). Data are presented as mean ± SD. ∗p < 0.05, ns, not significant, ordinary one-way ANOVA.
(D and E) Immunoblotting for indicated proteins in EBs on day 4. Differentiation of control (C68 and C42) or DS/CHD (D49 and D19) iPSCs was induced by the EB-based differentiation protocol, with or without 6 μM ChIR99021 from day 2 to day 4. A representative immunoblot is shown in D. Quantification of the active β-Catenin/total β-Catenin ratio and ISL1 expression is shown in E. Data are presented as mean ± SD. Each filled circle represents one independent experiment, with 3 experiments for each iPSC line. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001, ns, not significant, Ordinary one-way ANOVA.
(F) Time to the appearance of beating clusters in control or DS/CHD cultures post-induction of monolayer-based differentiation. Controls (C62 and C68) and DS/CHD (D7 and D49) cultures were treated with 6 or 10 μM CHIR99021 between day 0 and day 1. Each filled circle represents one independent experiment, with 3 experiments for each iPSC line. Data are presented as mean ± SD. ∗∗∗∗p < 0.0001, ns, not significant, ordinary one-way ANOVA.
(G) Representative traces of Ca2+ transients measured with the Ca2+ indicator GCaMP6f in control iPSC-CMs (C68), DS/CHD iPSC-CMs (D49) with 6 μM CHIR99021, and DS/CHD iPSC-CMs with 10 μM CHIR99021. Control and DS/CHD iPSCs were induced to differentiation using the monolayer protocol. Ca2+ imaging was acquired on day 13. Merged traces in the box are shown on right. Quantification of Ca2+ transient amplitude and frequency is shown in Figure S3.
(H) GO analysis of the overlapping down- and upregulated DEGs in DS/CHD-differentiated cells on day 7 and the rescue effects of restoration of the activity of the Wnt signaling. Three independent experiments were performed for each iPSC line (C62, C68, D7, and D49) under each condition for RNA-seq. FPKM ≥30 was set as cutoff to filter transcripts. Fold change ≥1.5 and p < 0.05 were set as criteria to determine DEGs.
(I and J) Immunoblotting for indicated proteins in cells differentiated from 3 control (C62, C68, and C42) and 3 DS/CHD (D7, D49, and D19) iPSC lines on day 3. Cells were treated with vehicle or 1 μM JAKi from day 0 to day 3. Representative immunoblots are shown in I. Protein quantification is shown in J. Data are presented as mean ± SD. Three independent experiments were performed for each iPSC line. Each filled circle represents one independent experiment for an individual iPSC line. ∗∗∗∗p < 0.0001, ns, not significant, ordinary one-way ANOVA.
(K and L) Immunoblotting for indicated proteins in cells differentiated from 3 control (C62, C68, and C42) and 3 DS/CHD (D7, D49, and D19) iPSC lines on day 3 using the monolayer-based differentiation protocol or in cells differentiated from 2 control (C68 and C42) and 2 DS/CHD (D49 and D19) iPSC lines on day 4 using the EB-based differentiation protocol. Monolayer-based differentiating cells were cultured with either 6 or 10 μM CHIR99021 from day 0 to day 1. EBs were treated with vehicle or 6 μM ChIR99021 from day 2 to day 4. Representative immunoblots are shown in K. Protein quantification is shown in L. Data are presented as mean ± SD. Three independent experiments were performed for each iPSC line. Each filled circle represents one independent experiment for an individual iPSC line. ∗p < 0.05, ∗∗∗∗p < 0.0001, ns, not significant, Ordinary one-way ANOVA.