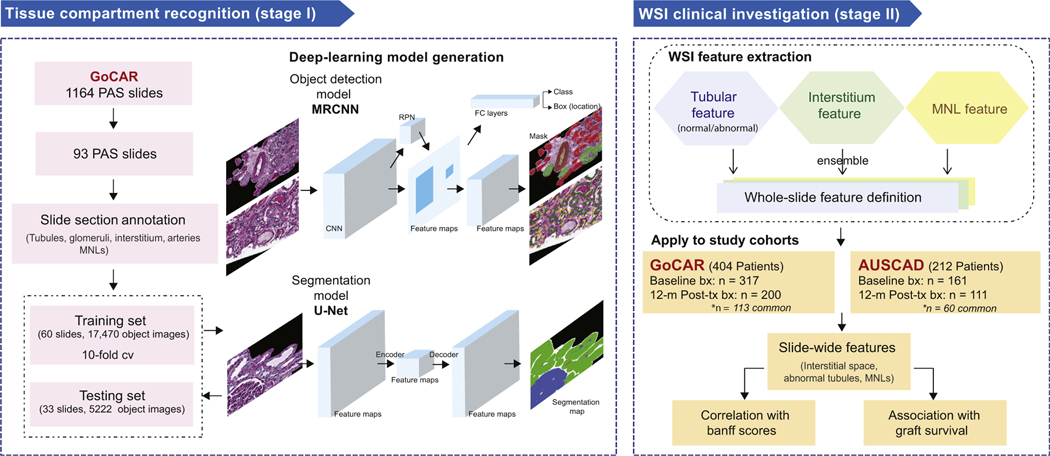
Figure 1 |. Study design.
This study consists of 2 major stages. Stage I is tissue compartment recognition. Ninety-three slides that represented the spectrum of histologic lesions were selected from the Genomics of Chronic Allograft Rejection (GoCAR) periodic acid–Schiff (PAS) slides and then randomly divided into the discovery set (n = 60) and the testing set (n = 33). The annotated sections of these slides were used for deep-learning model construction and evaluation. During the training process, we built the models based on 2 types of deep-learning structures for compartment or mononuclear leukocyte (MNL) detection (by mask region-base convolution neural network [MRCNN]) and tissue segmentation (by U-Net). Models were determined through evaluation with 10-fold cross-validation and finally applied to the testing set. Stage II is the whole-slide image (WSI) clinical investigation. Using the established deep-learning model, we processed 789 baseline and 12-month post-transplantation (post-tx) WSIs from 2 independent cohorts (GoCAR and Australian Chronic Allograft Dysfunction [AUSCAD]) and extracted a series of slide-wide digital features capturing the abnormalities in the interstitium and tubules, and MNL infiltration. These features were further examined through association with Banff scores and post-transplantation graft survival. bx, biopsies; FC, fully connected; RPN, region proposal network.