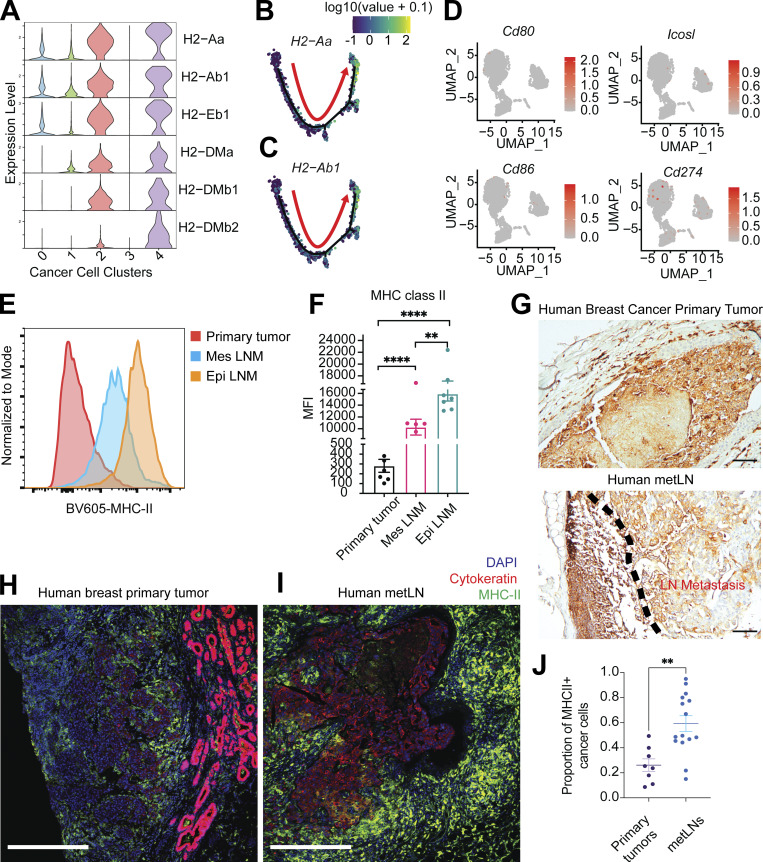
Figure 2.
The presence of MHC-II on cancer cells during LNM. (A) Violin plot represents the gene expression of MHC-II molecules in 4T1 cancer cells. (B and C) Expression of MHC-II molecules H2-Aa and H2-Ab1 were projected to the single-cell trajectories. Gene expression values are scaled and log-normalized. The red arrow indicates the pseudotime trajectory of cancer cells’ progression. (D) UMAP of single-cell gene expression of costimulatory and coinhibitory molecules in cancer cells. (E) Surface MHC-II presence on cancer cells measured by flow cytometry. (F) The mean fluorescent intensity (MFI) of MHC-II on cancer cells from 4T1 primary tumors and metLNs. Mes LNM, EpCAM−vimentin+; Epi LNM, EpCAM+vimentin−. One-way ANOVA was used for statistical analysis; **, P < 0.01; ****, P < 0.0001. Tukey’s multiple comparisons test was used for the post-hoc test. (G) Immunohistochemistry staining of MHC-II (HLA-D) in human breast cancer primary tumor and metLN. The scale bar represents 100 μm in all images. (H and I) Immunofluorescence staining of nuclei (DAPI, blue), pan-cytokeratin (red), and MHC-II (green) in human breast cancer primary tumor (H) and matched metLN (I). The thickness of the tissue section is 5 μm. The scale bar represents 500 μm in all images. (J) The proportion of MHC-II–positive cancer cells in human breast tumors (n = 8) and LNM (n = 15). Cell annotation was performed using QuPath software. **, P < 0.01. Student’s t test was used for the statistical analysis.