Figure 1. Dimer-forming p53(A347D) is hyperstable and transcriptionally impaired.
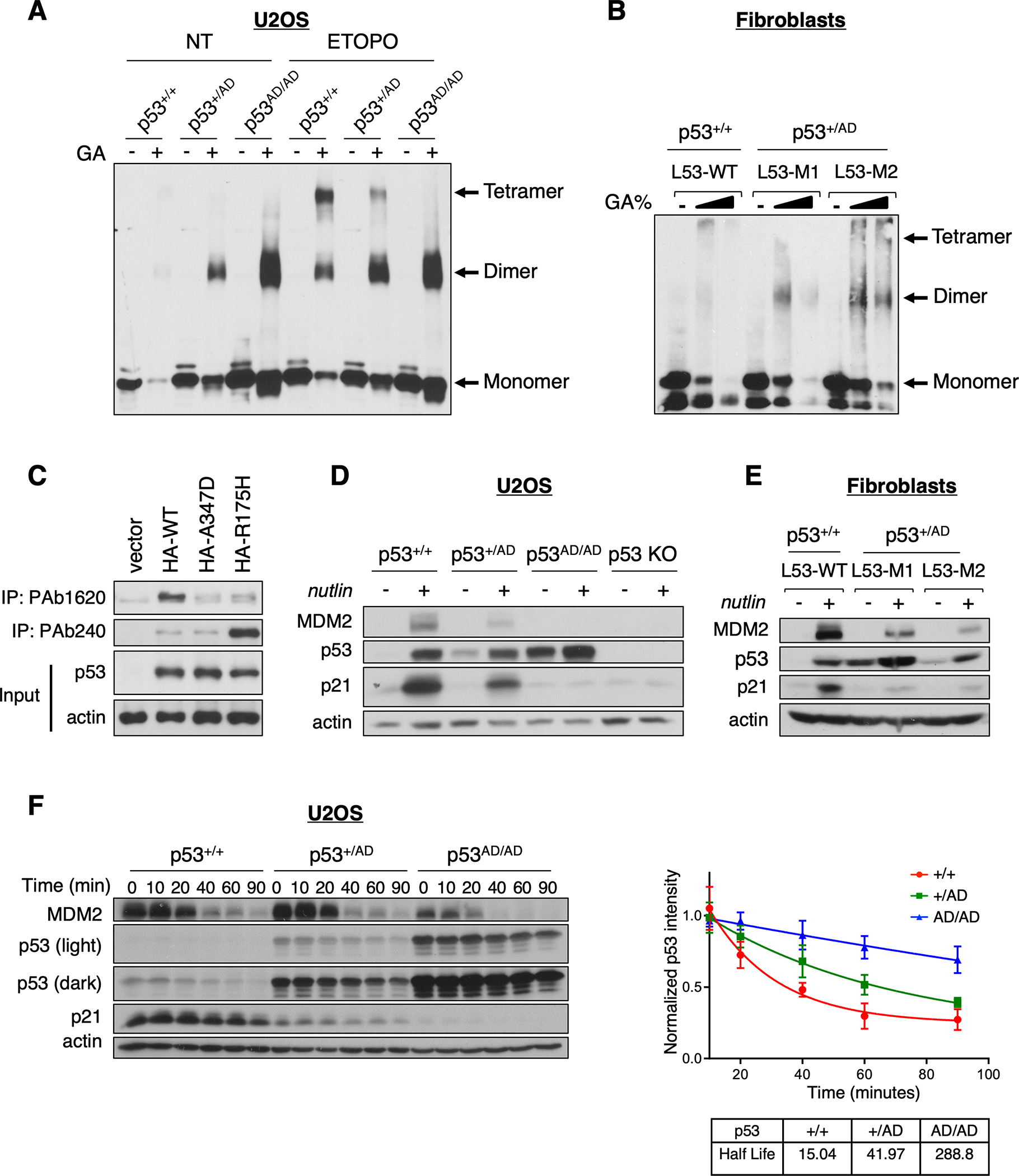
(A) Protein lysates from U2OS p53+/+, p53+/AD, and p53AD/AD cells treated with (ETOPO) or without (NT) 20 μM etoposide for 6 h were incubated in the presence or absence of 0.005% glutaraldehyde for 20 min at room temperature (RT) and subjected to immunoblot analysis with a monoclonal p53 antibody (DO1/1801) to detect p53 oligomeric species indicated at right.
(B) Protein lysates from primary dermal fibroblasts expressing WT p53 (L53-WT) or heterozygous p53(A347D) (L53-M1, L53-M2) were incubated with an increasing concentration of glutaraldehyde (0%, 0.01%, 0.05%) for 20 min at RT then subjected to immunoblot analysis as in A.
(C) U2OS p53 KO cells were transfected with plasmids expressing HA-WT-p53, HA-p53(A347D), HA-p53(R175H) or the empty vector pcDNA3. Protein lysates were subjected to immunoprecipitation with anti-p53 PAb240 or PAb1620 and immunoblot analysis with anti-p53 (DO-1).
(D) U2OS p53+/+, p53+/AD, p53AD/AD, and p53 KO cells were treated with 10 μM nutlin-3a for 24 h then lysed. Protein lysates were then subjected to immunoblot analysis with antibodies against indicated proteins.
(E) Protein lysates from primary dermal fibroblasts varying in p53 status were treated with 10 μM nutlin-3a for 24 h, and then processed for immunoblotting with antibodies against indicated proteins.
(F) (Left) Following the addition of cycloheximide (100 μg/mL), U2OS cells expressing p53+/+, p53+/AD, and p53AD/AD were harvested at the indicated times. Cell lysates were then subjected to immunoblotting. (Right) Densitometric analysis was performed using ImageJ to assess the half-life of p53. Each point represents the density of the p53 band at indicated time points relative to the initial time point. Data represent mean ± SEM for three biological replicates.
