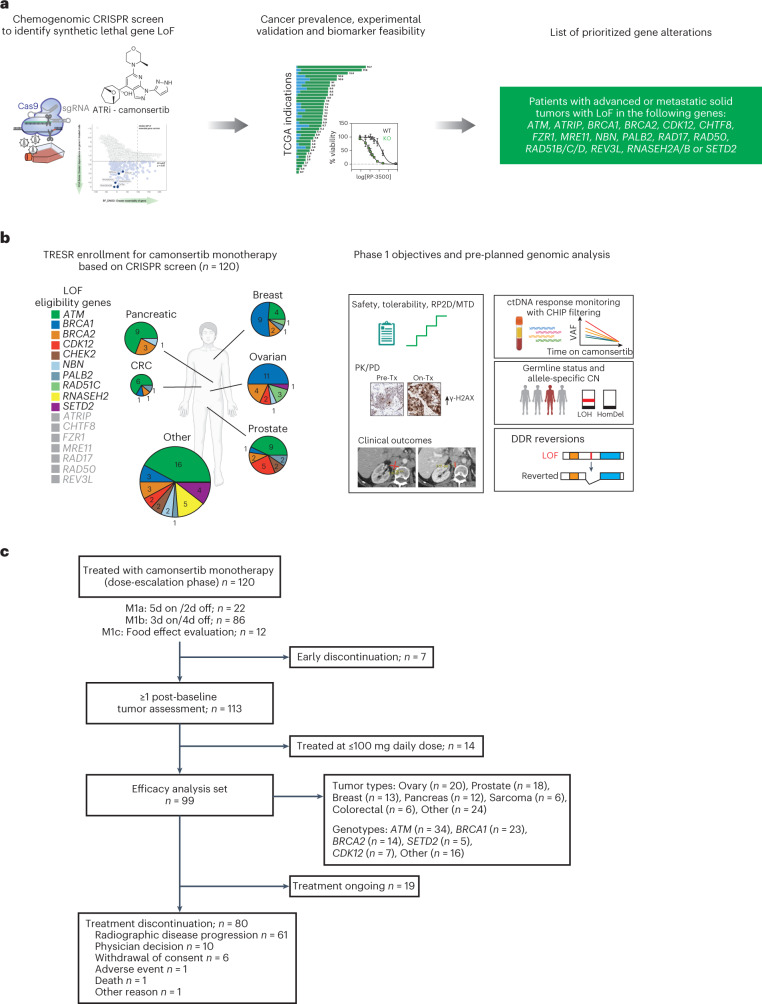
Fig. 1. Overview of the TRESR trial: a CRISPR–Cas9 chemogenomic-informed clinical trial.
a, SNIPRx CRISPR–Cas9-enabled chemogenomic screen to identify ATRi-sensitizing and synthetic lethal alterations for patient selection. b, Patient enrollment by gene and tumor type and overview of pre-planned analyses, which included (1) clinical endpoints; (2) PK in plasma and PD in pre-treatment and on-treatment biopsies; (3) hypothesis-generating genomic analyses, such as the assessment of allelic status (that is, biallelic versus non-biallelic alterations) and somatic versus germline status; and (4) analysis of longitudinal ctDNA as an early marker of camonsertib activity. c, CONSORT diagram of TRESR monotherapy patient populations. Patients enrolled in M1c received a single dose on day 3 in the fed state and continued from day 1 (fasted state) on either the 5/2 (n = 3) or 3/4 (n = 9) schedule. 3/4, 3 d on, 4 d off; 5/2, 5 d on, 2 d off; CN, copy number; CRC, colorectal cancer; HomDel, homozygous deletion; M, module; TCGA, The Cancer Genome Atlas; Tx, therapy.