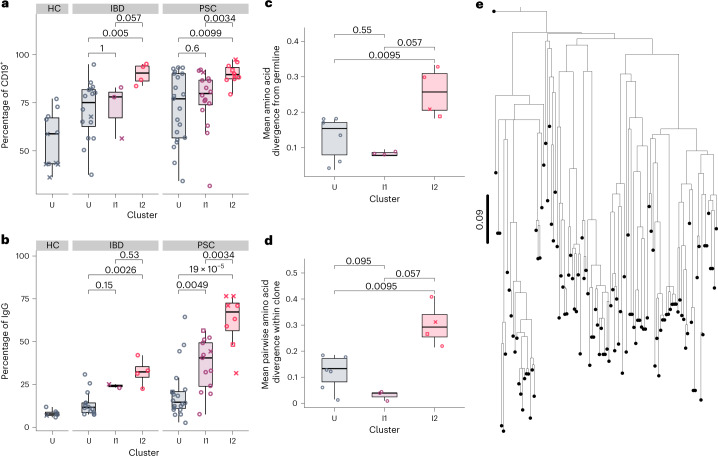
Fig. 4. PSC inflammation is characterized by an influx of IgG plasma cells and plasma cells show signs consistent with an antigen drive.
a, Proportion of right colon plasma cells positive for surface CD19 by flow cytometry (n = 10 PSC I2, n = 16 PSC I1, n = 21 PSC U, n = 4 IBD I2, n = 3 IBD I1, n = 16 IBD U, n = 11 HC U). b, Proportion of IgG-secreting plasma cells among total right colon plasma cells as determined by enzyme-linked immunosorbent spot (ELISpot) (n = 8 PSC I2, n = 13 PSC I1, n = 20 PSC U, n = 4 IBD I2, n = 2 IBD I1, n = 14 IBD U, n = 6 HC U). c, Mean amino acid divergence from the inferred germline within CDR3 of the largest clones identified in each patient (n = 4 PSC I2, n = 3 PSC I1, n = 6 PSC U). d, Mean pairwise amino acid divergence within CDR3 of the largest clones identified in each patient (n = 4 PSC I2, n = 3 PSC I1, n = 6 PSC U). Each symbol represents an individual patient (open circles denote patients without dysplasia at the time of sampling; ‘x’ denotes patients with dysplasia at the time of sampling; open squares denote patients indefinite for dysplasia at the time of sampling). The center line represents the median; the hinges indicate the first and third quartiles; the upper and lower whiskers extend to the largest and smallest values within 1.5 times the IQR from the first and third quartiles, respectively (a–d). e, Dendrogram of heavy chain sequences within the top clone of an I2 patient (n = 110 sequences). This clone demonstrates a ‘lop-sided’ branching pattern, which is consistent with nonrandom mutation accumulation and antigen drive. The origin point represents the inferred germline sequence. The scale bar represents the codon substitutions per codon.