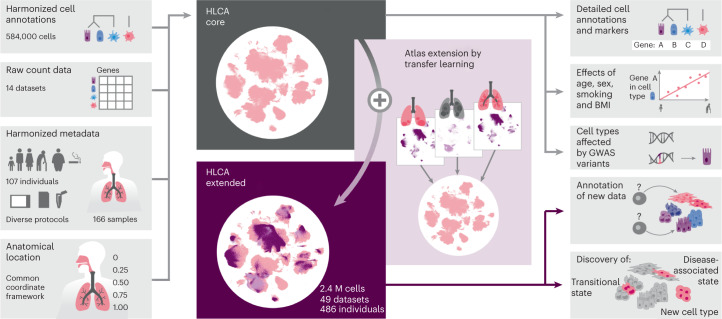
Fig. 1. HLCA study overview.
Harmonized cell annotations, raw count data, harmonized patient and sample metadata and sample anatomical locations encoded into a CCF were collected and generated as input for the HLCA core (left). After integration of the core datasets, the atlas was extended by mapping 35 additional datasets, including disease samples, to the HLCA core, bringing the total number of cells in the extended HLCA to 2.4 million (M). The HLCA core provides detailed consensus cell annotations with matched consensus cell type markers (top right), gene modules associated with technical, demographic and anatomical covariates in various cell types (middle right), GWAS-based association of lung conditions with cell types (middle right) and a reference projection model to annotate new data (middle right) and discover previously undescribed cell types, transitional cell states and disease-associated cell states (right, bottom).