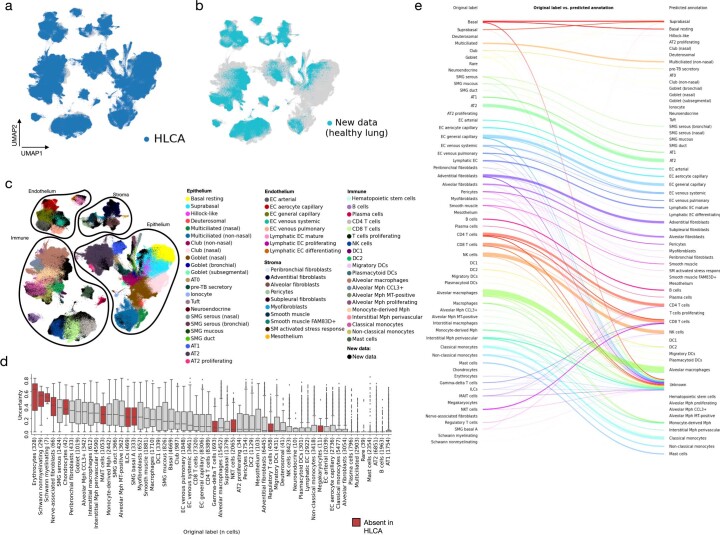
Extended Data Fig. 6. Mapping of unseen healthy lung scRNA-seq data to the HLCA core.
a, UMAP of the jointly embedded HLCA core (dark blue, plotted on top) and the newly mapped healthy lung data (gray). b, Same as a, but now plotting cells from the HLCA in gray, and cells from the new data on top in light blue. c, Same as a, but now coloring cells from the HLCA core by their final annotation, and coloring cells from the new data in black. Cells from each of the compartments are outlined to ease visual identification of cell types by colors. d, Uncertainty of label transfer (ranging from 0 to 1) for cells from the mapped data, subdivided by original cell type label. Number of cells per label is shown between brackets. Cell labels are ordered by mean uncertainty. Boxes of cell labels not present in the HLCA core are colored red. Boxes show median and interquartile range of uncertainty. Cells with uncertainties more than 1.5 times the interquartile range away from the high and low quartile are considered outliers and plotted as points. Whiskers extend to the furthest non-outlier point. e, Sankey plot of original labels of cells from the mapped dataset versus predicted annotations based on label transfer. Cells with uncertainty >0.3 are labeled ‘unknown’. AT: alveolar type. DC: dendritic cells. EC: endothelial cells. ILCs: innate lymphoid cells. MAIT cells: mucosal-associated invariant T cells. MT: metallothionein. Mph: macrophages. NK: natural killer. NKT cells: natural killer T cells. SM: smooth muscle. SMG: submucosal gland. TB: terminal bronchiole.