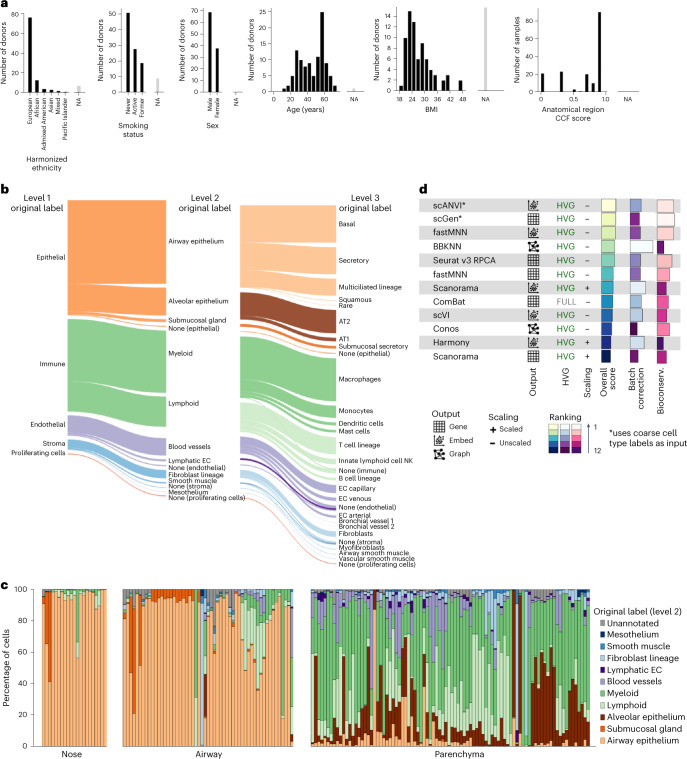
Fig. 2. Composition and construction of the HLCA core.
a, Donor and sample composition in the HLCA core for demographic and anatomical variables. Donors/samples without annotation are shown as not available (NA; gray bars) for each variable. For the anatomical region CCF score, 0 represents the most proximal part of the lung and airways (nose) and 1 represents the most distal (distal parenchyma). Donors show diversity in ethnicity (harmonized metadata proportions: 65% European, 14% African, 2% admixed American, 2% mixed, 2% Asian, 0.4% Pacific Islander and 14% unannotated; see Methods), smoking status (52% never, 16% former, 15% active and 17% NA), sex (60% male and 40% female), age (ranging from 10–76 years) and BMI (20–49; 30% NA). b, Overview of the HLCA core cell type composition for the first three levels of cell annotation, based on harmonized original labels. In the cell type hierarchy, the lowest level (1) consists of the coarsest possible annotations (that is, epithelial (48% of cells), immune (38%), endothelial (9%) and stromal (4%)). Higher levels (2–5) recursively break up coarser-level labels into finer ones (Methods). Cells were set to ‘none’ if no cell type label was available at the level. Cell labels making up less than 0.02% of all cells are not shown. Overall, 94, 66 and 7% of cells were annotated at levels 3, 4 and 5, respectively. c, Cell type composition per sample, based on level 2 labels. Samples are ordered by anatomical region CCF score. d, Summary of the dataset integration benchmarking results. Batch correction score and biological conservation score each show the mean across metrics of that type, as shown in Supplementary Fig. 1, with metric scores scaled to range from 0 to 1. Both Scanorama and fastMNN were benchmarked on two distinct outputs: the integrated gene expression matrix and integrated embedding (see output). The methods are ordered by overall score. For each method, the results are shown only for their best-performing data preprocessing. Methods marked with an asterisk use coarse cell type labels as input. Preprocessing is specified under HVG (that is, whether or not genes were subsetted to the 2,000 (HVG) or 6,000 (FULL) most highly variable genes before integration) and scaling (whether genes were left unscaled or scaled to have a mean of 0 and a standard deviation of 1 across all cells). EC, endothelial cell; NK, natural killer; Bioconserv., conservation of biological signal.