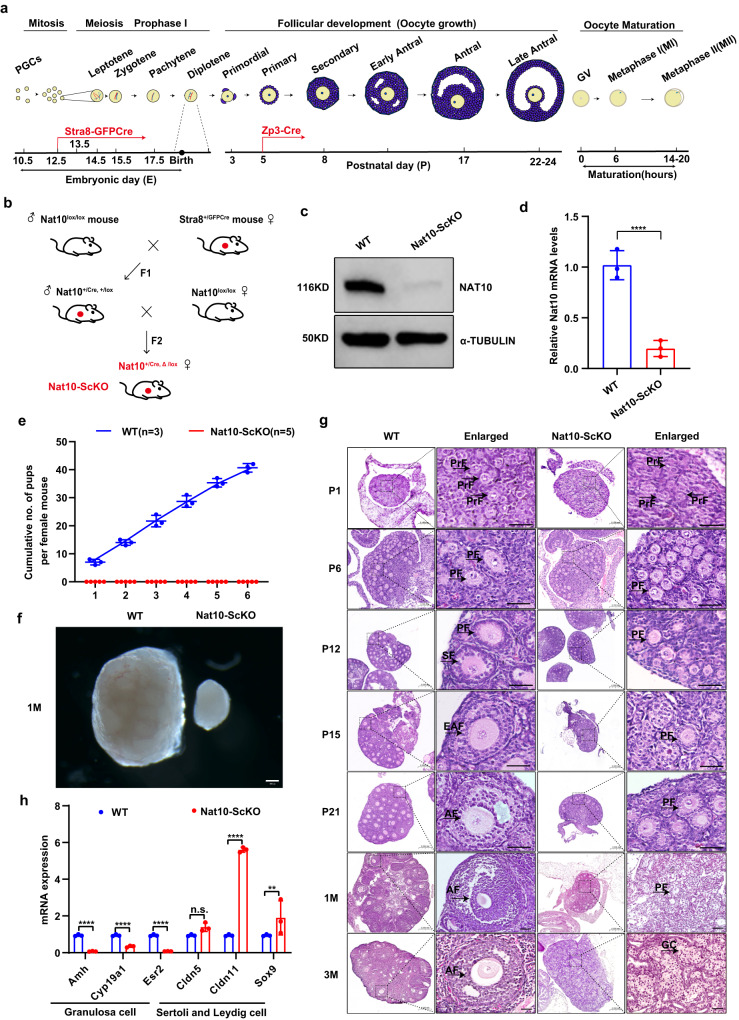
Fig. 2. Pre-meiotic deletion of Nat10 caused follicular developmental arrest and premature ovarian failure (POF).
a Schematic diagram showing the landmark timeline of oocyte development from embryonic meiotic cell-cycle progression to postnatal oocyte growth and maturation. Stra8-GFPcre is activated prior to Embryonic day 13.5 (E13.5); Zp3-cre is active starting from P5 in the primary follicles. b A breeding scheme by crossing Nat10lox/lox with Stra8-GFPCre to generate Nat10lox/-; Stra8-GFPCre (Nat10-ScKO) offspring. c Western blot analyses of the NAT10 protein levels in adult WT and Nat10-ScKO ovary. α-TUBULIN was used as a loading control. d Quantitative RT–PCR (qPCR) assay showing the relative expression levels of Nat10 mRNA in adult WT and Nat10-ScKO mouse ovary. Data are presented as mean ± SEM, n = 3. ****p < 0.0001 by two-tailed Student’s t-test. e Fertility test showing the cumulative average numbers of pups from breeding of WT and Nat10-ScKO females with WT males. Data are presented as the mean ± SEM, n = 5, ****p < 0.0001 by two-tailed Student’s t-test. f The gross morphology of ovaries derived from WT and Nat10-ScKO mice at 1 M. Scale bar, 200 μm. n = 3 biologically independent samples were included in each group. g H&E staining of paraffin-embedded ovarian sections showing the histology of WT and Nat10-ScKO ovaries at postnatal days as indicated. Scale bar, 200 μm. A high-resolution view of the boxed area is shown in parallel. Scale bar, 20 μm. n = 3 biologically independent samples were included in each group. Arrows point to follicles at stages as indicated. PrF primordial follicle, PF primary follicle, SF secondary follicle; EAF early antral follicle, AF antral follicle, LAF late antral follicle. h qPCR analyses of the relative expression levels for a cohort of genes showing specific or characteristic expression in ovarian granulosa cells (Left) or testicular Sertoli/Leydig cells (Right) in 1-month-old WT and Nat10-ScKO ovaries. Data are presented as the mean ± SEM, n = 3. n.s., not significant; **p < 0.01; ****p < 0.0001 by two-tailed Student’s t-test. Cldn5, p = 0.3326; Sox9, p = 0.0059. n = 3 biologically independent samples were included in each group (c, f, g). Source data are provided as a source data file.