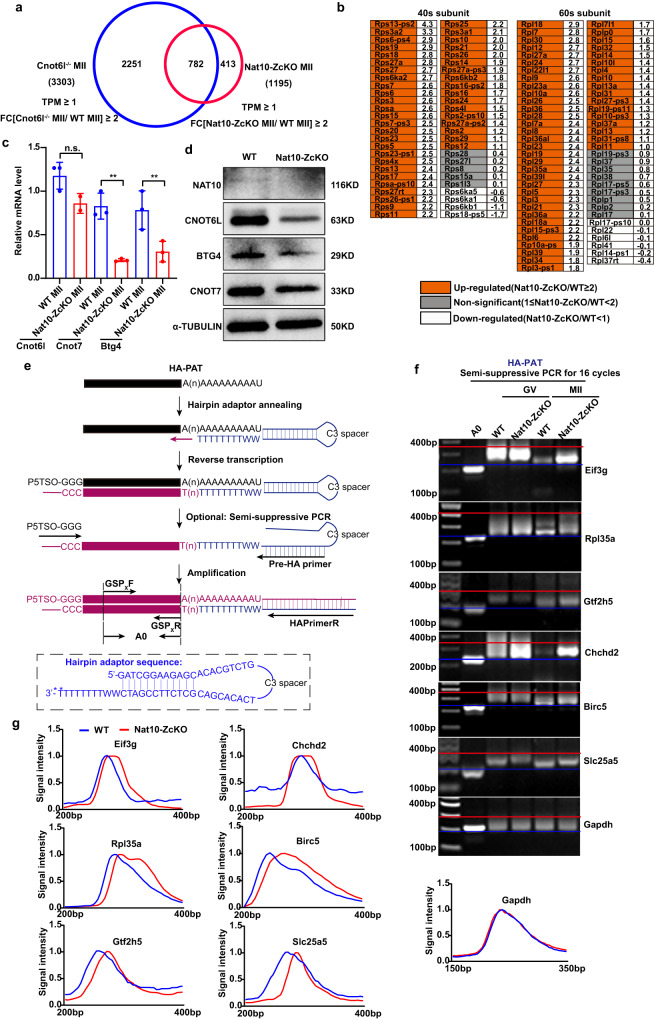
Fig. 8. Hairpin Adaptor-Poly(A) Tail length (HA-PAT) assay validated the deficient maternal mRNA decay in Nat10-ZcKO MII oocytes.
a Venn diagram showing the overlapping of transcripts that were stabilized during GV-to-MII transition in Cnot6l−/− and Nat10-ZcKO MII oocytes (FC = [WT MII/Nat10-ZcKO MII]≥2, p < 0.05). b Fold change of relative expression levels of transcripts encoding ribosomal protein subunits in Nat10-ZcKO relative to WT oocytes at the MII stage. The values of log2(FC[Nat10-ZcKO/WT]) are listed in the right column. c qPCR results showing the relative levels of indicated transcripts (Cnot6l, Cnot7 and Btg4) in WT and Nat10-ZcKO oocytes at MII stage. Data are presented as the mean ± SEM, n = 3. n.s., non-significant, **p < 0.01 by two-tailed Student’s t-test. Cnot6l, p = 0.0906; Cnot7 p = 0.0020; Btg4 p = 0.0031. d Western blot displaying the NAT10, CNOT6L, CNOT7 and BTG4 protein levels in MII oocytes of WT and Nat10-ZcKO mice. α-TUBULIN was used as a loading control. n = 3 biologically independent samples were included in each group. e A schematic illustration depicting the design strategy and the key steps for Hairpin Adaptor-Poly(A) Tail length (HA-PAT) assay. The 1st strand of cDNA was synthesized with the hairpin adaptor (HA) primer in conjunction with a P5TSO primer containing three “G”, via a mechanism of “template-switching”. GSP, Gene-specific primer; A0, the PCR product resulting from the amplification with a gene-specific pair of GSPxF and GSPxR primers; GSPxR primer was designed against an mRNA’s 3′ terminals preceding the poly(A) sequence; poly(A)-containing PCR products were amplified with GSPxF and fixed HAPrimerR primers. The full sequence for hairpin adaptor (HA) is listed at the bottom. W indicates degenerate nucleotides (A or T); * The asterisk indicates the phosphorothioate modification. f, g HA-PAT assay results showing changes in poly(A)-tail lengths of indicated transcripts in WT and Nat10-ZcKO oocytes at GV and MII stages. Experiments were performed in triplicates; a representative image is shown in the 2% agarose gel in f and the length distribution is shown in the densitometric curves in (g). n = 3 biologically independent samples were included in each group (d, f). Source data are provided as a source data file.