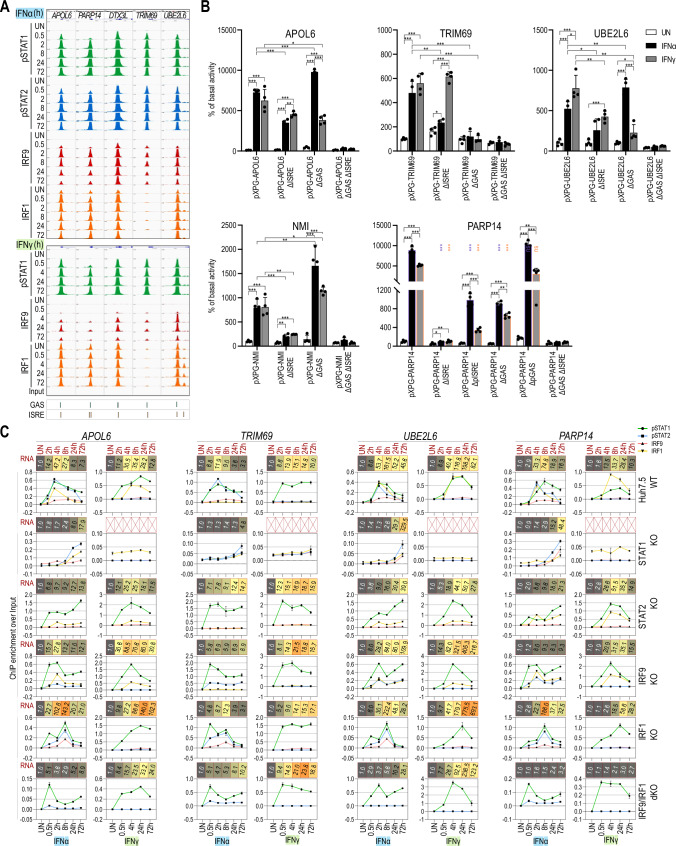
Fig. 5.
The ISRE + GAS composite site shares features of GAS and ISRE genes and acts as a molecular switch in response to IFNα and IFNγ. A Representative views of the ChIP-seq peaks detected in the promoter regions of APOL6, PARP14, DTX3L, TRIM69 and UBE2L6 genes, containing GAS + ISRE binding sites, in untreated or treated with IFNα or IFNγ Huh7.5 cells. All peaks were mapped onto human reference genome hg38 and visualized using the IGV genome browser; scale for STATs 0–1000, IRFs 0–500; p phosphorylated. B Results of luciferase-based reporter assay for APOL6, TRIM69, UBE2L6, NMI and PARP14 promoters. Wild-type and mutated (Δ)GAS and/or ISRE sequences of the promoter regions were cloned into pXPG vector. Huh7.5 cells were co-transfected with pXPG (Firefly luciferase—FLUC) and pRL-SV40 (Renilla luciferase—RLUC) expression vectors and left untreated or stimulated with IFNα or IFNγ for 8 h. After harvesting, cells were lysed and the fluorescence of FLUC and RLUC levels were measured. Bars indicate the Relative Luminescence Units (RLU) of the sample calculated as a percentage of basal activity of the untreated sample transfected with pXPG vector containing wild-type sequence of the promoter. n = 3; mean ± SEM, p calculated with unpaired two-tailed Student t test, *p < 0.05, **p < 0.01, and ***p < 0.001; To maintain clarity, we denoted statistical significance between the response of wild-type and mutated constructs in our PARP14 experiments using violet and orange stars for IFNα- and IFNγ-treated samples, respectively (graph shows the representative experiment). C Detailed characteristics of APOL6, PARP14, TRIM69 and UBE2L6 genes. Line charts indicated ChIP-qPCR assays performed on APOL6, PARP14, TRIM69 and UBE2L6 gene promoters in Huh7.5 wild-type or knock-out cell lines treated with IFNα or IFNγ (n = 2, mean ± SEM). Red boxes represent expression data from RNA-seq experiment (n = 2; mean fold change over untreated sample). RNA-seq experiment for Huh7.5 STAT1KO treated with IFNγ was not performed