Abstract
Background
Aquaporin 9 (AQP9) is permeable to water or other small molecules, and plays an important role in various cancers. We previously found that AQP9 was related to the efficacy of chemotherapy in patients with colorectal cancer (CRC). This study aimed to identify the role and regulatory mechanism of AQP9 in CRC metastasis.
Methods
The clinical significance of AQP9 was analysed by using bioinformatics and tissue microarray. Transcriptome sequencing, Dual-Luciferase Reporter Assay, Biacore, and co-immunoprecipitation were employed to demonstrate the regulatory mechanism of AQP9 in CRC. The relationship between AQP9 and CRC metastasis was verified in vitro and in vivo by using real-time cell analysis assay, high content screening, and liver metastasis models of nude mice.
Results
We found that AQP9 was highly expressed in metastatic CRC. AQP9 overexpression reduced cell roundness and enhanced cell motility in CRC. We further showed that AQP9 interacted with Dishevelled 2 (DVL2) via the C-terminal SVIM motif, resulting in DVL2 stabilization and the Wnt/β-catenin pathway activation. Additionally, we identified the E3 ligase neural precursor cell expressed developmentally downregulated 4-like (NEDD4L) as a modulator regulating the ubiquitination and degradation of AQP9.
Conclusions
Collectively, our study revealed the important role of AQP9 in regulating DVL2 stabilization and Wnt/β-catenin signaling to promote CRC metastasis. Targeting the NEDD4L–AQP9–DVL2 axis might have therapeutic usefulness in metastatic CRC treatment.
Keywords: aquaporin 9, colorectal cancer metastasis, Dishevelled 2, Wnt/β-catenin signaling
Graphical abstract
Graphical Abstract.
Schematic diagram of AQP9 promoting CRC metastases.
Introduction
Colorectal cancer (CRC) is now the third most common cancer worldwide [1]. About 50% patients with CRC initially diagnosed with localized disease will eventually progress to metastatic disease [2]. Distant metastases are the major cause of CRC-related death [3, 4]. The development of antibodies to epidermal growth factor receptor (EGFR) was a milestone in metastatic CRC treatment. However, the EGFR-targeted therapy is only recommended to treat patients with KRAS wild-type (WT) tumors. Metastatic CRC remains a therapeutic challenge and is considered a non-curable disease.
The role of Wnt signaling in CRC tumorigenesis, metastasis, and poor patient survival has been well documented [5]. Extensive efforts have been made to block Wnt signaling, but always with poor results [6]. Dishevelled (DVL) is the hub of Wnt signaling transduction [7]. It has been demonstrated that DVL can interact with a wide range of proteins via its Postsynaptic density 95, Discs Large, Zonula occludens-1 (PDZ) domain, thus transmitting Wnt signaling from receptors to downstream effectors. Therefore, unveiling the DVL-associated protein interaction may provide valuable information for developing new CRC therapeutic intervention [8].
Aquaglyceroporins (AQPs) are a family of integral membrane proteins that are permeable to water and other small molecules. There is compelling evidence that aberrant AQPs expression correlated with tumorigenesis and cancer metastasis [9–11], fostering a growing interest in identifying AQPs as diagnostic and therapeutic targets in cancers. We previously found that aquaporin 9 (AQP9) was overexpressed in CRC tissues and its expression level was positively correlated with chemotherapy efficacy [12]. As the role and mechanism of AQP9 in CRC have not been fully elucidated, herein, we further investigated the possible role in CRC metastasis. Furthermore, structural and functional studies highlighted the importance of understanding the post-translational modifications and protein–protein interactions in regulating AQPs function [13]. Given the presence of a putative PDZ-binding motif in the C-terminal of AQP9 [14], we postulated that AQP9 might interact with the PDZ protein DVL2 and be involved in the regulation of Wnt signaling.
In the present study, we found that a high expression level of AQP9 in CRC was correlated with increased risks of metastasis and poor prognosis. AQP9 is important to maintain the epithelial–mesenchymal transition (EMT) state and the motility and metastasis of CRC cells. Mechanistically, AQP9 could interact with DVL2 via its C-terminal SVIM motif and subsequently affected Wnt signaling transduction. In addition, we identified the E3-ubiquitin ligase neural precursor cell expressed developmentally downregulated 4-like (NEDD4L) as a modulator of AQP9.
Materials and methods
Patients and samples
Tissue microarrays were constructed using paraffin-embedded samples of patients with primary colorectal adenocarcinomas at the Sixth Affiliated Hospital of Sun Yat-sen University (SYSU; Guangzhou, Guangdong, China) from 1 January 2007 to 31 December 2012. Patient enrollment criteria included: pathological confirmation of colorectal adenocarcinoma, the undergoing of curative surgical resection, absence of preoperative or post-operative chemotherapy, availability of tumor specimen, and complete follow-up information. The median follow-up time was 1,490 days (range, 195–2,636 days). Overall survival (OS) or progression-free survival (PFS) was the end point of the study. OS time was calculated from the date of surgery to the date of death or the last follow-up time. PFS time was calculated from the date of surgery to the date of disease progression.
Written informed consent from each patient regarding tissue sampling had been obtained and the study was reviewed and approved by the Medical Ethics Committee of the Sixth Affiliated Hospital of SYSU (2020ZSLYEC-236).
Plasmid constructs, small interfering RNAs (siRNAs), and transfections
The pcDNA 3.1 (+) plasmid was purchased from Invitrogen (Carlsbad, CA, USA). The pCI–HA–NEDD4L plasmid was obtained from Addgene (Cambridge, MA, USA). For AQP9 transient transfection, pEGFP or pEGFP–AQP9 plasmids (Addgene) were transfected into CRC cells by using Lipofectamine® 3000(Invitrogen). siRNAs (Ribobio; Guangzhou, Guangdong, China) were transfected into CRC cells by using Lipofectamine® RNAiMAX (Invitrogen).
For stable transfection, full length of human AQP9 cDNA open reading frame was cloned into the lentiviral expression vector pCDH–CMV–MCS–EF1–copGFP (SBI Pharmaceuticals; Tokyo, Japan). HEK293T cells were co-transfected with pCDH, pCMV–Δ8.91, and pCMV–VSVG plasmids (Addgene) to produce lentivirus. Luciferase-expressing HCT116 cells were infected with collected virus. Infected cells were treated with polybrene (Biosharp; Hefei, Anhui, China) and selected with puromycin (Selleck; Houston, TX, USA) for 2 weeks. All the plasmid constructions were confirmed by sequencing.
Cell culture and immunofluorescence
CRC cell lines were purchased from the American Type Culture Collection (Gaithersburg, MD, USA). HCT116 and DLD1 cells were maintained in RPMI-1640 (Gibco; Carlsbad, CA, USA), supplemented with 10% (v/v) fetal bovine serum (Gibco). Cells were allowed to grow in a humidified incubator with 5% CO2 at 37°C. Mycoplasmas were detected in cell cultures stained with 4’,6-diamidino-2-phenylindole (DAPI) (Santa Cruz; Dallas, TX, USA).
Cells were fixed using 4% paraformaldehyde for 20 min at 4°C. Fixed cells were then permeabilized in 0.5% Triton X-100 for 10 min and blocked with 5% bovine serum albumin for 1 h. Primary antibody incubation was performed at 4°C overnight. The cell nuclei were counterstained with DAPI. The images were analysed using the SP8 confocal microscope (Leica; Wetzlar, Germany).
Co-immunoprecipitation and Western blot analysis
For the co-immunoprecipitation (Co-IP) assay, cells were plated in a 100-mm dish and transfected with the indicated plasmids. After 48 h of transfection, cells were incubated with 500 μL of Pierce™ IP Lysis Buffer (ThermoFisher; Carlsbad, CA, USA). Then, 350 μL of whole-cell lysates were incubated with the indicated antibody at 4°C overnight and 30 μL of protein G-Sepharose beads were added at 4°C for 8 h. The beads were washed five times using IP wash buffer and precipitates were eluted using 30 μL of 2X sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS–PAGE) sample buffer and analysed by using Western blot with the indicated antibody.
For the nuclear and cytoplasm separation, total proteins were extracted from the cultured cells using RIPA Lysis and Extraction Buffer (ThermoFisher). Nuclear and cytoplasm proteins were extracted using a KeyGene KIT (QIAGEN; Hilden, Germany). The proteins separated on SDS–PAGE were transferred onto a polyvinylidene fluoride membrane (Millipore; Darmstadt, Germany). The membrane was blocked for 1 h in Tris-Buffered saline and Tween 20 (TBST; 10 mM Tris-Cl, 150 mM NaCl, and 0.05% Tween 20) containing 4% bovine serum albumin. All primary antibodies were incubated overnight at 4°C. Horseradish peroxidase (HRP)-linked anti-rabbit or mouse IgG secondary antibodies (ThermoFisher) were used to detect primary antibody binding. Protein was detected by using the Electrochemiluminescence system (Millipore) on autoradiography film (Kodak; Rochester, NY, USA) or ChemiDoc Touch (Bio-rad; Hercules, CA, USA).
The following antibodies were used: anti-actin (Proteintech; Rosemont, IL, USA) or anti-GAPDH (Proteintech) as loading controls; anti-catenin, anti-E-cadherin, anti-Snail, anti-DVL2, and anti-GFP (Proteintech); anti-AQP9 or anti-ubiquitination (Santa Cruz); anti-HA (Ser139), anti-Flag, and anti-NEDD4L (Cell Signaling Technology; Danvers, MA, USA).
Real-time cell growth curve assay
For cells monitored by using the xCELLigence real-time cell analysis assay DP instrument (Roche; Basel, Switzerland), 5,000 cells per well were seeded on 16-well cell invasion and migration plates (Roche). Cell activities are expressed as the cell impedance index.
Luciferase assay
CRC cells seeded in 24-well plates were co-transfected with pGL4.49[luc2P/TCF-LEF RE/Hygro] Vector (Promega; Madison, WI, USA) and one of these plasmids (pEGFP, pEGFP–AQP9–WT, pEGFP–AQP9–△SVIM). At 48 h after transfection, luciferase activity was measured using the Dual-Luciferase Reporter Assay (Promega). Experiments were performed in five replicates and the means with standard errors were calculated. Data were normalized with an internal control, pGL4.74[hRluc/TK] Vector (Promega).
Protein docking simulation with deep-learning models
The sequences of AQP9 (O43315) and DVL2 (O14641) were obtained from The Universal Protein Resource [15]. At the same time, the sequence of the DVL2PDZ domain was obtained from the Conserved Domain Database [16]. Then, SWISS-MODEL [17] and trRosetta [18] were used to build the protein models according to the sequences of AQP9 and the DVL2PDZ domain. The AQP9 protein model was built by using trRosetta with restraints from both deep-learning and homologous templates, where the template modeling score was used to evaluate the model [19]. The DVL2PDZ domain was established by using homology modeling in the SWISS-MODEL sever and the modeling result was evaluated using the MolProbity Score [20].
Then we used the “prepare protein” module [21, 22] in Discovery Studio 2017R2 to optimize the structure of the two proteins, including removing crystal water and treating disulfide bonds, treating metal ions, and adding terminal hydrogen atoms to protein molecules according to the expected temperature and pH value. Afterwards, the ZDOCK module was employed to predict the connection between AQP9 and the DVL2PDZ domain. Before running, the Euler angle step size of the ligand direction of rotation sampling was set to 5, “RMSD Cutoff” was set to 6.0, “Interface Cutoff” was set to 9.0, “Maximum Number of Clusters” was set to 60, and the combination mode “Top Poses” was set to 2,000, with the default values for other parameters. The RDOCK module was applied to obtain the composite of the docking pose with the highest score. The “Analyze Protein Interface” module was utilized to analyse the connection between AQP9 and the DVL2PDZ domain.
Protein purification
For AQP9 C-terminal purification, a cDNA fragment (729–885 bp) corresponding to the C-terminal portion (containing the last transmembrane domain) of human AQP9 was amplified and cloned into the pGEX-4T-1 expression vector. Purified AQP9-C-terminal protein was added with an N-terminal glutathione S-transferase (GST) tag (GST–AQP9-C-term). The SVIM motif was deleted (GST–AQP9–△SVIM) to obtain a truncated protein. For protein expression, different constructs were transformed into the BL21 (DE3) competence cell. The expression was induced by using isopropyl β-D-thiogalactoside and evaluated by using SDS–PAGE. The bacteria were cultured and the protein was purified by using GST affinity chromatography followed by a molecule sieve. For DVL2 purification, full length of the DVL2 gene was amplified and cloned into the pET30a expression vector. The BL21 bacteria were amplified and the protein was purified by using Ni-IDA affinity chromatography. Supernatants were aliquoted and stored at −80°C.
Biacore assay
For the prebinding experiment, pure AQP9 and AQP9–△SVIM proteins were diluted using 10-mM sodium acetate buffers in a range of different pH values (pH 3.0–5.5) and flowed across the surface of the chip at a rate of 10 μL/min. Absorption of the protein to the chip was observed and the subsequent CM5 chip coupling was selected by combining the most suitable binding concentration.
For CM5 chip coupling, a certain volume of AQP9 and AQP9–△SVIM protein was diluted using sodium acetate buffer to 20 μg/mL at pH 5.0 according to the preconjugation ratio. Three reagents, N-hydroxysuccinimidobiotin (NHS), 1-(3-Dimethylaminopropyl)-3-ethylcarbodiimide hydrochloride (EDC), and ethanolamine hydrochloride, were equilibrated at room temperature and placed on the rack of a Biacore T200 (GE; Atlanta, GA, USA). The liquid flow rate was set to 10 μL/min. The chip flowed through 125 μL of NHS/EDC mixture, 166 μL of sample, and 140 μL of ethanolamine hydrochloride, successively.
For conjugation, pure DVL2 protein was diluted to the appropriate concentration (0, 75, 150, 300, 600, and 1,200 μg/mL) using Phosphate buffered saline-P20) buffer (Cytiva; Freiburg, BW, Germany (PBS-P+) and loaded to observe binding activity. The binding dissociation curve was obtained at different protein concentrations. For chip regeneration, the chip was flushed using 10 mmol/L of glycine (pH 2.5) to restore the baseline to initial values. The results were then analysed.
Murine CRC model of liver metastasis
Female BALB/c nude mice aged 4–6 weeks were purchased from the Model Animal Research Center of Nanjing University (Nanjing, China). HCT116 cells were engineered to express luciferase by using lentiviral transfection of pEZ–Lv105 plasmid (Addgene). The CDs region of the AQP9 was amplified by using polymerase chain reaction (PCR) and cloned into the pCDH–CMV–MCS–EF1–copGFP–T2A–Puro vector (Addgene). HCT116-Luc cells were then transfected with the pCDH control vector or pCDH–AQP9 vector. To model liver metastasis, 500,000 cells were suspended in 50 μL of PBS and injected into the spleen. All surgical procedures were performed under isoflurane anesthesia. Tumor growth and metastatic spread were assessed weekly via an In Vivo Imaging System (PerkinElmer; Waltham, MA, USA). The present studies in animals were reviewed and approved by the Animal Care and Use Committee of SYSU (IACUC–2020090901).
Statistics and reproducibility
All experiments were performed at least in triplicate. Statistical analyses were performed using GraphPad Prism 8.0 (La Jolla, CA, USA) and SPSS 22.0 (Chicago, IL, USA). Statistical analyses for cell-line experiments were performed by using Student’s t-test or Mann–Whitney U test. Data are presented as mean ± standard error of the mean (SEM) with each dot representing an individual experiment. For in vivo assays in xenografts, statistical analyses were performed by using two-way analysis of variance (ANOVA). A P-value of <0.05 was considered statistically significant (*P < 0.05, **P < 0.01, ***P < 0.001).
OS time was calculated from the date of surgery to the date of death or the last follow-up time if the follow-up time was >5 years. For PFS, an event was defined as the first clinical or pathologic evidence of local or distant recurrence or death. Receiver operation characteristic curve analysis was applied to determine the cut-off point for tumor “high expression” by using the 0, 1-criterion. Under this condition, a score value of 4.8 was adopted as the cut-off for stratification of AQP9 expression into low (≥4.8) and high (<4.8). A score value of 4.3 was adopted as the cut-off for stratification of DVL2 expression into low (>3.84) and high (≤3.84). The relationship between AQP9 and clinicopathologic features of patients with CRC was analysed by using chi-square test. The Kaplan–Meier method was used for the univariate survival analysis and the differences between compared groups were assessed by using the log-rank test. The Cox proportional hazards regression model was used to compare the OS and PFS between marker categories and to obtain risk ratios.
Availability of data and materials
All data generated or analysed in this study are included in this published article. All the data supporting the findings of this study are available from the corresponding author upon request.
Results
AQP9 upregulation is correlated with CRC progression and metastasis
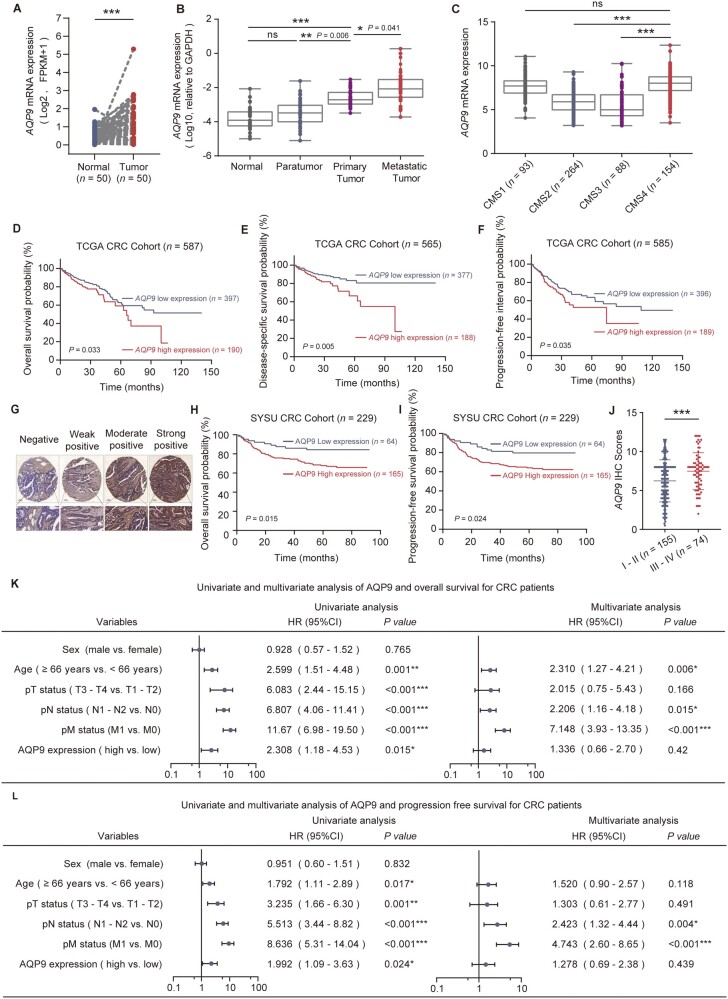
Using The Cancer Genome Atlas (TCGA) data from 50 tumor-normal paired CRC samples and 698 unpaired CRC samples, we found that AQP9 expression level was significantly higher in tumor samples than in normal samples and it had certain accuracy in predicting tumor occurrence (Figure 1A andSupplementary Figure 1A and B). Elevated AQP9 expression was observed in later-stage CRC compared with stage I CRC (Supplementary Figure 1C). Consistently, our immunohistochemical (IHC) analysis showed a strong staining signal of AQP9 in CRC tumor tissues, implicating an oncogenic role of AQP9 (Supplementary Figure 1D). Furthermore, we compared AQP9 mRNA levels in matched primary tumors, metastatic tumors, adjacent normal tissues, and paratumor tissues of 39 patients with CRC and distant metastases; 33 of 39 primary tumors had substantially elevated levels of AQP9 with respect to adjacent normal tissues and paratumor tissues (Supplementary Figure 1E) and AQP9 expression in metastatic tumors was considerably higher than that in primary tumors and paired normal tissues or paratumor tissues (Figure 1B). These data suggested that elevation of AQP9 might be related to tumor progression and metastases. Consensus molecular subtype (CMS) is a CRC classification system based on molecular alterations such as genomic drivers, transcriptomic subtypes, and immune signatures [23, 24]. Interestingly, we found that a high AQP9 level was significantly associated with the “mesenchymal” CMS4 CRC (Figure 1C), indicative of high invasion ability and poor survival [24]. In parallel, AQP9 high expression was significantly associated with poor prognosis (Figure 1D), especially a lower probability of disease-specific survival (Figure 1E) and progression-free interval (Figure 1F), indicating that AQP9 might play an important role in CRC progression and metastasis.
Figure 1.
AQP9 high expression is associated with CRC progression and metastasis. (A) AQP9 mRNA expression in TCGA–CRC tumor tissues compared with matched normal tissues (n = 50, P < 0.001), paired two-tailed t-test. (B) AQP9 mRNA level determined by using RT–qPCR in paired normal tissues, paratumor tissues, tumor, and metastatic tumors (n = 39). (C) AQP9 mRNA level in TCGA–CRC tumors separated by using CMS. Kaplan–Meier plots of (D) OS, (E) disease-specific survival, (F) progression-free interval layered by AQP9 mRNA level (TCGA data sets, log-rank test). (G) Representative images of immunohistochemistry staining of AQP9 in SYSU CRC tissue microarray. (H) OS and (I) PFS of patients with CRC (SYSU-cohort) stratified by using AQP9 IHC score. (J) Quantification of AQP9 IHC score in patients with CRC separated by Stage I–II and Stage III–IV. Univariate and multivariate analyses for (K) OS and (L) PFS. All data are shown as mean ± SEM. *P < 0.05; **P < 0.01; ***P < 0.001. AQP9, aquaporin 9; CRC, colorectal cancer; TCGA, The Cancer Genome Atlas; CMS, consensus molecular subtypes; OS, overall survival; PFS, progression-free survival; SYSU, Sun Yat-sen University; IHC, immunohistochemical; T, tumor; N, node; M, metastasis; SEM, standard error of the mean; ns, no significance.
To further validate the clinical significance and interaction of AQP9, we subjected the tumor tissue microarray in 229 patients with CRC by using IHC staining (Figure 1G). Kaplan–Meier analysis showed that a higher expression level of AQP9 was associated with a lower probability of OS (P = 0.015) (Figure 1H) and PFS (P = 0.024) (Figure 1I) than a lower expression level of AQP9 in patients with CRC. A higher AQP9 IHC score was significantly associated with a higher tumor, node, and metastasis (TNM) stage (Figure 1J). Though multivariate Cox regression analysis showed that AQP9 was not an independent predictor of OS and PFS for patients with CRC (Figure 1K and L), we found that AQP9 expression levels were significantly associated with sex, median age, clinical stage, disease progression, or metastasis in patients with CRC (Table 1).
Table 1.
Association between expression of AQP9 and clinicopathological features of patients with CRC
| Variable | Low AQP9 | High AQP9 | P-value |
|---|---|---|---|
| Sex | 0.077 | ||
| Male | 42 (65.6%) | 87 (52.7%) | |
| Female | 22 (34.4%) | 78 (47.3%) | |
| Median age (years) | 0.026* | ||
| <66 | 38 (59.4%) | 71 (43.0%) | |
| ≥66 | 26 (40.6%) | 94 (57.0%) | |
| Clinical stage | 0.002** | ||
| I–II | 53 (82.8%) | 102 (61.8%) | |
| III–IV | 11 (17.2%) | 63 (38.2%) | |
| pT status | 0.088 | ||
| T1–T2 | 24 (37.5%) | 43 (26.1%) | |
| T3–T4 | 40 (62.5%) | 122 (73.9%) | |
| pN status | 0.008** | ||
| N0 | 53 (82.8%) | 107 (64.8%) | |
| N1–N2 | 11 (17.2%) | 58 (35.2%) | |
| pM status | 0.094 | ||
| M0 | 59 (92.2%) | 138 (83.6%) | |
| M1 | 5 (7.8%) | 27 (16.4%) | |
| Disease progression | 0.016* | ||
| No | 51 (79.7%) | 104 (63.0%) | |
| Yes | 13 (20.3%) | 61 (37.0%) |
P < 0.05;
P < 0.01;
P < 0.001.
CRC, colorectal cancer; T, tumor; N, node; M, metastasis.
AQP9 sustains cell migration and invasion, and is associated with EMT
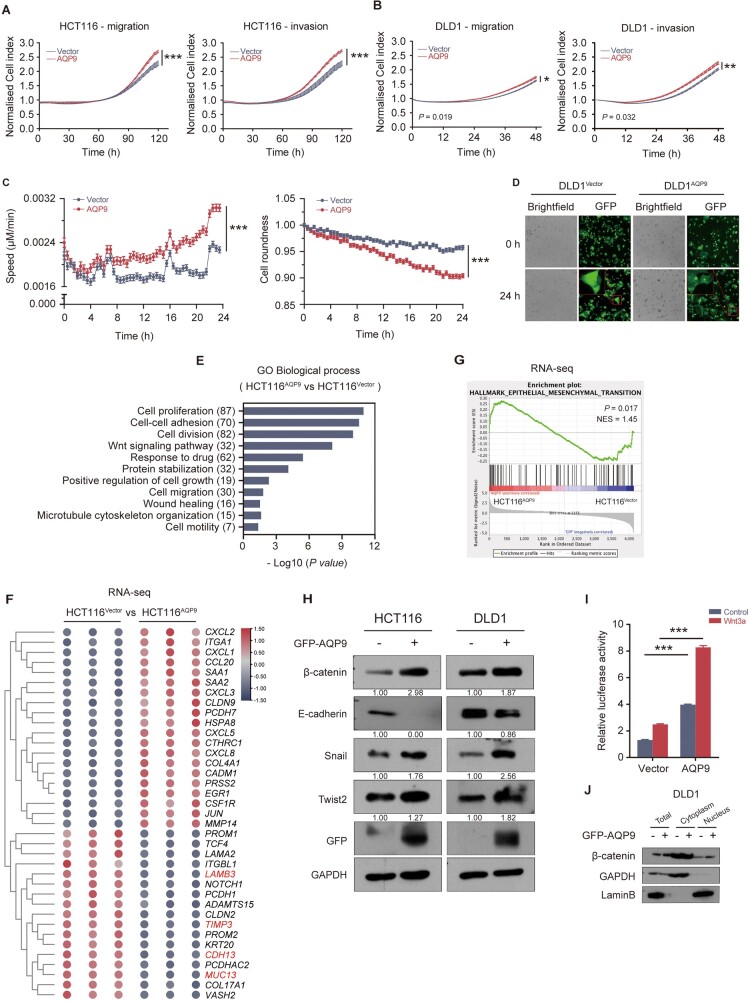
Given that AQP9 was enriched in CMS4 tumors, which had significant overexpression proteins implicated in mesenchymal activation and high invasion ability, we next investigated whether AQP9 had an impact on cell migration and EMT. As expected, AQP9 overexpression significantly promoted the migration and invasion of CRC cells (Figure 2A and B). High content screening showed that AQP9 resulted in increased cell movement speed and reduced cell roundness (Figure 2C andSupplementary Figure 2A). Compared with the control cells, AQP9-overexpressed cells showed a significant projected area of lamellar morphology, indicative of a migratory and invasive phenotype (Figure 2D and Supplementary Figure 2B).
Figure 2.
AQP9 overexpression promotes migration and invasion in CRC cells and regulating the Wnt/β-catenin pathway. (A) and (B) CRC cell migration and invasion monitored by using real-time cell analysis assay. (C) Cell movement speed and cell roundness measurement by using high content screening. (D) Cell morphology of control cells and AQP9-overexpressed cells. Scale bar represents 100 μm. (E) GO-biological process analysis on RNA-seq data of HCT116AQP9 vs HCT116Vector. (F) Heat map of cell adhesion genes in HCT116AQP9 and HCT116Vector cells. (G) GSEA analysis using our RNA-seq data. (H) Western blot analysis of EMT markers including β-catenin, E-cadherin, Snail, and Twist2 in HCT116 and DLD1 cells after transfection with AQP9-overexpressed plasmids. (I) Statistics of TCF/LEF reporter luciferase assay in HCT116 cells. (J) Western blot of cytoplasmic/nucleoplasmic experiment of CRC cells transiently expressing AQP9. All data are shown as mean ± SEM. *P < 0.05; **P < 0.01; ***P < 0.001. GO, gene ontology; RNA-seq, RNA sequencing; AQP9, aquaporin 9; CRC, colorectal cancer; EMT, epithelial–mesenchymal transition; TCF/LEF, T-cell factor/lymphoid enhancer factor; GSEA, gene set enrichment analysis; SEM, standard error of the mean; ns, no significance.
Then we performed gene expression analysis by using Ribonucleic acid (RNA) sequencing (RNA-seq) to examine the molecular characteristics of AQP9-overexpressed CRC cells. We identified 255 significantly deregulated genes relative to control cells (72 up, 183 down, ≥ 2-fold, P-value < 0.05) (Supplementary Figure 2C). Gene Ontology (GO) term analysis was calculated using the DAVID online tool (https://david.ncifcrf.gov/summary.jsp; Supplementary Figure 2D). Consistently with the cell phenotype, cell attachment and motility-related gene sets were significantly enriched among the top GO terms (Figure 2E). Clustering of the expression profile for cell-migration-related genes is shown in Figure 2F. In AQP9-overexpressed cells, most cell adhesion genes were downregulated (e.g. MUC13, CDH13, TIMP3, and LAMB3), whereas cell detachment-associated genes such as chemokines, MMP14, and PCDH7 were upregulated. Further gene set enrichment analyses (GSEAs) using our RNA-seq data revealed that AQP9 high expression was significantly associated with enrichment of gene sets in EMT (Figure 2G), indicating the involvement of AQP9 in regulating EMT-associated cell movement. We specifically analysed the positive correlation between AQP9 gene expression and EMT in a TCGA–CRC data set (Supplementary Figure 2E). Multiple data sets highlighted the positive correlation between AQP9 and EMT (Supplementary Figure 2F). In agreement with the RNA-seq results, Western blot showed that AQP9 overexpression could affect the expression of EMT markers, including upregulation of β-catenin, Snail, and Twist2, and downregulation of E-cadherin (Figure 2H).
Many previous studies showed Wnt/β-catenin signaling activation and nuclear β-catenin increase during EMT [5, 25]. In the nucleus, β-catenin associates with T-cell factor/lymphoid enhancer factor (TCF/LEF) transcription factors and drives the transcription of downstream oncogenes, which can subsequently enhance EMT progression. As GEO data sets analysis also indicated the enrichment of Wnt signaling (Supplementary Figure 2G), we therefore tried to determine whether AQP9-induced EMT is associated with Wnt/β-catenin signaling activation. We performed a TCF/LEF reporter luciferase assay using HCT116 cells transfected with AQP9 together with TCF/LEF–Luc. AQP9 overexpression significantly enhanced the transcriptional activity of the luciferase reporter, which was more obvious when activated by using Wnt3a (Figure 2I), suggesting activation of the canonical Wnt pathway. Moreover, AQP9 overexpression significantly enhanced levels of nuclear β-catenin (Figure 2J). Collectively, these results demonstrated that AQP9 induced a migratory phenotype via the Wnt/β-catenin pathway in CRC cells.
AQP9 modulates the Wnt/β-catenin pathway in CRC through binding to and stabilizing DVL2
The Wnt/β-catenin pathway is activated when a Wnt ligand binds to the transmembrane Frizzled (FZD) receptor and its co-receptor and then recruits the scaffolding protein DVL to the cell membrane to initiate a downstream Wnt signal cascade [26]. DVL2 is a PDZ protein that contains a highly conserved PDZ domain to stabilize its membrane localization [7, 27]. Membrane proteins such as receptors and channels usually form functional clusters by directly binding to PDZ proteins [28, 29]. Previous study revealed that the binding cleft of the DVLPDZ domain was more flexible than those of canonical PDZ domains and enabled recognition of both C-terminal and internal peptides [8]. Given the presence of a PDZ-binding motif SVIM at the C-terminal cytoplasmic tail of AQP9 [14] (Figure 3A), we postulated that AQP9 might interact with DVL2 and be involved in the regulation of Wnt signaling. Therefore, we generated a predicted binding model of the AQP9–DVL2PDZ complex via deep-learning-based methods. The confidence of the predicted AQP9 model was very high with an estimated template modeling score of 0.842 and the modeling result of the DVL2PDZ domain was good with a MolProbity Score of 0.74. One hundred and forty-six docking poses were first generated by using the ZDOCK program and the conformation of the AQP9 C-terminal with the highest score was selected for further optimization using the RDOCK program (Figure 3B). It could be seen that the two proteins binding interfaces produced eight hydrogen bonds and two conjugated π bonds (Table 2).
Figure 3.
AQP9 stabilizes the DVL2 protein via the C-terminal SVIM motif. (A) C-terminal peptide of the PDZ domain structure of AQP9 and DVL2. (B) Deep-learning-based AI prediction of AQP9 and DVL2 binding. (C) and (D) Co-IP experiment to confirm the interaction between AQP9 and DVL2. Co-IP experiment of CRC cells co-transfected with DVL2, (E) AQP9–△cterm, and (F) AQP9–△SVIM (F). (G) Immunofluorescence of AQP9 localization after transfection with AQP9–WT, AQP9–△cterm, and AQP9–△SVIM plasmids. (H) Biacore binding affinities analysis. All data are shown as mean ± SEM. *P < 0.05; **P < 0.01; ***P < 0.001. AQP9, aquaporin 9; CRC, colorectal cancer; WT, wild-type; SEM, standard error of the mean; ns, no significance.
Table 2.
The interaction between residues at the docking interface of AQP9 and the DVL2PDZ domain
| AQP9 residue | DVL2 residue | Distance/Å | Type |
|---|---|---|---|
| VAL293 | LEU14 | 2.0689 | H-bond |
| GLU283 | ASN23 | 2.1771 | H-bond |
| GLU283 | ASN66 | 2.6429 | H-bond |
| VAL293 | GLY15 | 2.7046 | H-bond |
| GLU287 | ARG74 | 2.8475 | H-bond |
| LYS288 | ILE18 | 2.8691 | H-bond |
| MET295 | LEU14 | 2.9636 | H-bond |
| SER292 | ILE16 | 3.2834 | H-bond |
| MET295 | HIS78 | 5.8522 | π-bond |
| ILE294 | PHE13 | 5.2412 | π-bond |
P < 0.05;
P < 0.01;
P < 0.001.
Å, angstroms.
Hydrogen bonds, especially short and strong hydrogen bonds, play extremely important roles in stabilizing the structure of proteins [30, 31]. Among the eight hydrogen bonds, AQP9:VAL293:O-DVL2:LEU14:HN was the most stable with a distance of 2.0689 angstroms, indicating that the binding site was quite stable. Residues SER292, VAL293, and MET295 of the SVIM motif also produced strong hydrogen bonds. It is worth noting that MET295 also produced a sulfur–π interaction, while ILE294 produced an alkyl–π interaction. The above results indicated that DVL2 could stably bind to the C-terminal end of the AQP9 protein via hydrogen bond and conjugated π bond interaction force.
To confirm the predicted binding between AQP9 and DVL2, we first performed Co-IP assays and found that both endogenous and exogenous DVL2 could interact with AQP9 in CRC cells, respectively (Figure 3C and D). To further verify the predicted binding site, we then constructed truncated AQP9 plasmids, in which the last 30 amino acids of the C-terminal (AQP9–△cterm) or SVIM motif (AQP9–△SVIM) had been deleted. Co-IP assays showed that deletion of the C-terminal or SVIM motif affected the interaction between AQP9 and DVL2 (Figure 3E and F). Interestingly, the confocal microscopy experiments demonstrated that the overexpression of AQP9–△cterm or AQP9–△SVIM plasmids resulted in the membrane-to-cytoplasm translocation of AQP9 (Figure 3G), suggesting that the C-terminal, especially the SVIM motif, was important for AQP9 membrane localization and might be necessary for the interaction with DVL2. To address the contribution of the AQP9–SVIM motif to the direct binding with DVL2, we purified GST–AQP9 fusion proteins corresponding to either the C-terminal (GST–AQP9–cterm–WT) or a truncated C-terminal in which the SVIM motif had been deleted (GST–AQP9–cterm–△SVIM). Biacore SPR analysis showed that AQP9–cterm–WT bound to DVL2 with higher affinity compared with the AQP9–cterm–△SVIM mutants (Figure 3H). The reduced binding affinity and DVL2 protein level in the AQP9–SVIM mutants were consistent with Co-IP experimental results.
As we proved that AQP9 could directly bind to DVL2, it remained to clarify how AQP9 was regulating the activation of downstream Wnt/β-catenin signaling. Western blot showed that AQP9 overexpression resulted in a protein level increase in the DVL2 (Figure 4A), whereas deletion of the C-terminal or SVIM motif in AQP9 was associated with a decreased DVL2 level in CRC cells (Figure 4B). GSEA revealed an enrichment of gene sets for degradation of DVL in AQP9-overexpressed CRC cells (Figure 4C). Because the mRNA level of DVL2 was not changed by AQP9 overexpression (Supplementary Figure 3A), we speculated that AQP9 might upregulate the DVL2 level via regulating the stability of the DVL2 protein. To examine whether AQP9 could regulate the DVL2 protein turnover in CRC, we then performed cycloheximide assays and found that AQP9 could enhance the half-life of endogenous DVL2 and β-catenin compared with the controls (Figure 4D andSupplementary Figure 3B). Moreover, DVL2 knock-down could suppress the AQP9-induced β-catenin elevation in CRC cells (Figure 4E), suggesting that AQP9 might stabilize the DVL2 protein and subsequently inhibit downstream β-catenin degradation. Furthermore, TCF/LEF reporter luciferase assay showed that truncated AQP9 protein eliminated the transcriptional activity of the luciferase reporter compared with AQP9–WT proteins (Figure 4F). Taken together, the above results demonstrated that the AQP9 could bind to and stabilize DVL2, which subsequently activated the Wnt/β-catenin pathway.
Figure 4.
AQP9 activates the Wnt/β-catenin pathway via stabilization with DVL2. (A) Representative immunoblots of DVL2 protein level upon AQP9 overexpression. (B) Western blot shows DVL2 and β-catenin protein level. (C) GSEA analysis of DVL2 genes using HCT116 cell RNA-seq data. (D) CHX chase assays to determine the half-life of β-catenin and DVL2 in AQP9-overexpressed CRC cells. (E) Representative immunoblots of DVL2 and β-catenin protein level after being co-transfected with AQP9 plasmids and siDVL2. (F) Statistics of TCF/LEF reporter luciferase assay indicate that AQP9–cterm–△SVIM mutants eliminate the transcriptional activity of the luciferase reporter in HCT116 cells. All data are shown as mean ± SEM. *P < 0.05; **P < 0.01; ***P < 0.001. CHX, cycloheximide; RNA-seq, RNA sequencing; AQP9, aquaporin 9; CRC, colorectal cancer; TCF/LEF, T cell factor/Lymphoid enhancer factor; GSEA, gene set enrichment analysis; SEM, standard error of the mean; ns, no significance.
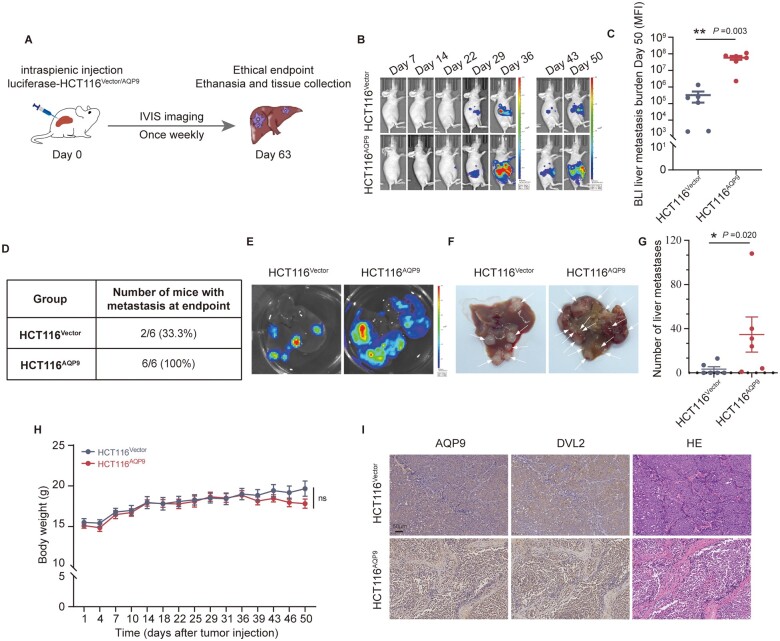
Expressing AQP9 upregulation promotes tumor metastasis in vivo
To test whether AQP9 promotes tumor metastasis in vivo, we cloned the AQP9 cDNA fragment into a lentiviral vector for stable infection and expression in mammalian cells. AQP9-overexpressed HCT116 cells (HCT116AQP9) or control cells (HCT116Vector) were injected into the spleen of nude mice (Figure 5A and Supplementary Figure 4A–D). The HCT116AQP9 group showed increased metastatic spread as demonstrated by a stronger bioluminescence signal in vivo (Figure 5B and C). Moreover, HCT116AQP9 cells had a higher liver metastatic rate and more metastatic foci in the liver than HCT116Vector cells (Figure 5D–G). The average weight of the mice was not significantly different in both groups (Figure 5H). IHC staining of mice liver tissue showed that AQP9 overexpression was correlated with enhanced DVL2 expression in the tumor (Figure 5I). The results above illustrated that AQP9 could promote CRC metastasis in vivo.
Figure 5.
AQP9-overexpressed cancer cells drive liver metastasis in vivo. (A) Schematic representation of intrasplenic injection of luciferase–HCT116 (HCT116Vector and HCT116AQP9) and In Vivo Imaging System monitoring of liver metastasis. (B) The average bioluminescence signals and (C) liver metastatic burden were determined by using an In Vivo Imaging System (HCT116Vector and HCT116AQP9, n = 6). (D) Statistics of metastatic number. (E)–(G) Representative images and statistics of metastatic foci (pointed out with arrow) in mice liver. (H) Body weight curves of mice in two groups. (I) IHC staining of AQP9, DVL2, and H & E staining in liver metastatic loci. All data are shown as mean ± SEM. *P < 0.05; **P < 0.01; ***P < 0.001. AQP9, aquaporin 9; BLI, bioluminescence; IHC, immunohistochemical; H & E, hematoxylin and eosin stain; SEM, standard error of the mean; ns, no significance.
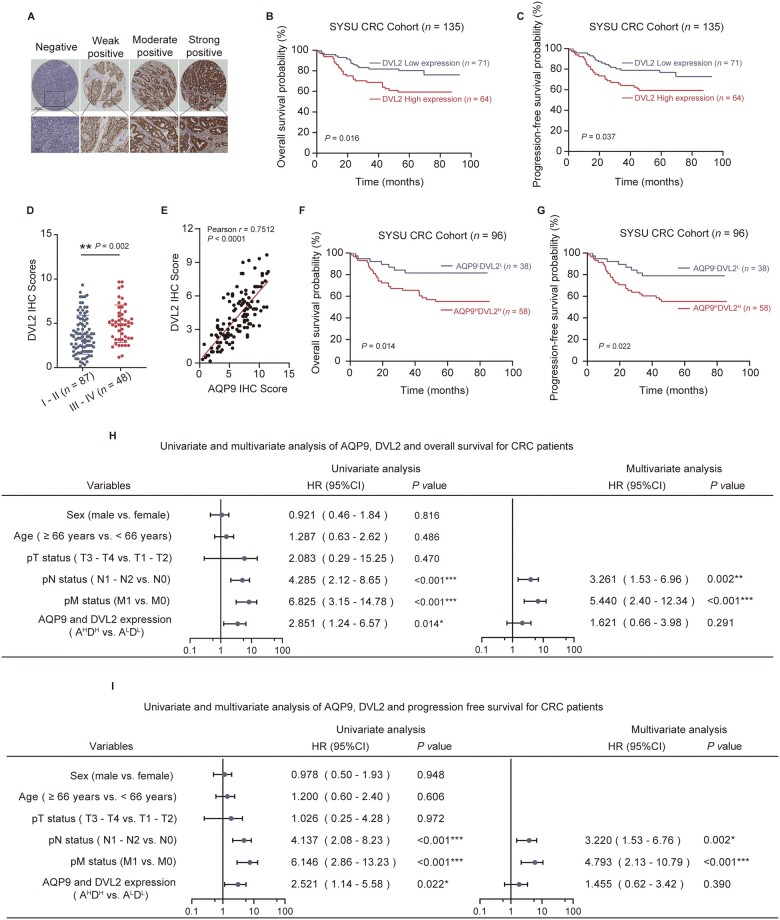
AQP9 and DVL2 expression is positively correlated with poor survival outcomes in patients with CRC
To confirm that AQP9 upregulation is correlated with upregulated DVL2 in patients with CRC, we performed IHC staining to analyse DVL2 expression using the tumor tissue microarray of 135 patients with CRC (Figure 6A). The DVL2 expression level was positively correlated with the clinical stage and pN status in patients with CRC (Supplementary Figure 5A and B, and Table 3). We also found that DVL2 high expression was associated with a low probability of OS (P = 0.016) and PFS (P = 0.037) (Figure 6B and C). A higher DVL2 IHC score was significantly associated with a higher TNM stage (Figure 6D). The IHC score indicated a strong (r = 0.7512) and significant (P < 0.001) positive association between AQP9 and DVL2 (Figure 6E). Most importantly, Kaplan–Meier analysis of combined AQP9 and DVL2 indicated that AQP9HighDVL2High patients with CRC had the lowest OS and PFS rate compared with AQP9LowDVL2Low patients with CRC (Figure 6F and G). High AQP9 and DVL2 expression was also significantly associated with clinical stage and pN status (Table 4).
Figure 6.
AQP9 and DVL2 high expression is correlated with poor outcome in CRC. (A) Representative images of IHC staining of DVL2 in SYSU CRC tissue microarray. (B) OS and (C) PFS of patients with CRC (SYSU-cohort), stratified by DVL2 IHC score. (D) Quantification of DVL2 IHC score in patients with CRC separated by Stage I–II and Stage III–IV. (E) Correlation analysis between AQP9 and DVL2 IHC score of patients with CRC (SYSU-cohort, n = 96, Pearson’s correlation, P < 0.001). (F) OS and (G) PFS of patients with CRC layered by the co-expression of AQP9 and DVL2 in tissue microarray. Univariate and multivariate analyses for (H) OS and (I) PFS. All data are shown as mean ± SEM. *P < 0.05; **P < 0.01; ***P < 0.001. AQP9, aquaporin 9; CRC, colorectal cancer; OS, overall survival; PFS, progression-free survival; SYSU, Sun Yat-sen University; IHC, immunohistochemical; T, tumor; N, node; M, metastasis; SEM, standard error of the mean; ns, no significance.
Table 3.
Correlation between expression of DVL2 and clinicopathological features of patients with CRC
| Variable | Low DVL2 | High DVL2 | P-value |
|---|---|---|---|
| Sex | 0.161 | ||
| Male | 44 (62.0%) | 32 (50.0%) | |
| Female | 27 (38.0%) | 32 (50.0%) | |
| Median age (years) | 0.129 | ||
| <66 | 30 (42.3%) | 19 (29.7%) | |
| ≥66 | 41 (57.7%) | 45 (70.3%) | |
| Clinical stage | 0.003** | ||
| I–II | 54 (76.1%) | 33 (51.6%) | |
| III–IV | 17 (23.9%) | 31 (48.4%) | |
| pT status | 0.123 | ||
| T1–T2 | 5 (7.0%) | 1 (1.6%) | |
| T3–T4 | 66 (93.0%) | 63 (98.4%) | |
| pN status | 0.003** | ||
| N0 | 57 (80.3%) | 36 (56.3%) | |
| N1–N2 | 14 (19.7%) | 28 (43.8%) | |
| pM status | 0.057 | ||
| M0 | 67 (94.4%) | 54 (84.4%) | |
| M1 | 4 (5.6%) | 10 (15.6%) | |
| Disease progression | 0.038* | ||
| No | 54 (76.1%) | 38 (59.4%) | |
| Yes | 17 (23.9%) | 26 (40.6%) |
P < 0.05;
P < 0.01;
P < 0.001.
CRC, colorectal cancer; T, tumor; N, node; M, metastasis.
Table 4.
Correlation between expression of AQP9 and DVL2, and clinicopathological features of patients with CRC
| Variable | AQP9LDVL2L | AQP9HDVL2H | P-value |
|---|---|---|---|
| Sex | 0.035* | ||
| Male | 26 (68.4%) | 27 (46.6%) | |
| Female | 12 (31.6%) | 31 (53.4%) | |
| Median age (years) | 0.112 | ||
| <66 | 20 (52.6%) | 21 (36.2%) | |
| ≥66 | 18 (47.4%) | 37 (63.8%) | |
| Clinical stage | <0.001*** | ||
| I–II | 32 (84.2%) | 28 (48.3%) | |
| III–IV | 6 (15.8%) | 30 (51.7%) | |
| pT status | 0.058 | ||
| T1–T2 | 4 (10.5%) | 1 (1.7%) | |
| T3–T4 | 34 (89.5%) | 57 (98.3%) | |
| pN status | 0.002** | ||
| N0 | 32 (84.2%) | 31 (53.4%) | |
| N1–N2 | 6 (15.8%) | 27 (46.6%) | |
| pM status | 0.083 | ||
| M0 | 36 (94.7%) | 48 (82.8%) | |
| M1 | 2 (5.3%) | 10 (17.2%) | |
| Disease progression | 0.017* | ||
| No | 30 (78.9%) | 32 (55.2%) | |
| Yes | 8 (21.1%) | 26 (44.8%) |
P < 0.05;
P < 0.01;
P < 0.001.
CRC, colorectal cancer; T, tumor; N, node; M, metastasis.
By using univariate and multivariate analysis, we found that the AQP9 and DVL2 expression level was not significantly associated with the OS and PFS rates, but was a strong, significant indicator of pN status (Figure 6H and I). Together, these results verified that the AQP9–DVL2 axis was upregulated in CRC and the expressions of AQP9 and DVL2 might be used as predictive biomarkers for the prognosis of patients with CRC.
The E3 ubiquitin ligase NEDD4L targets AQP9 for its degradation
Since the AQP9 level is an important determinant of CRC metastasis, we sought to identify AQP9-interacting E3 ligases that could regulate its protein stability. The E3 ligase NEDD4L caught our attention as a candidate since it has been shown to target several membrane proteins for proteasome degradation, including ion channels, growth factor receptors, and tight junction molecules [32–35]. The tryptophan, W (WW) domains of NEDD4L mediate its binding to substrates containing L/PPXY (PY) motifs [36]. The C-terminal of AQP9 contained a PY motif (LPFY), which was highly conserved among aquaglyceroporins (Figure 7A); this raised the possibility that AQP9 might be recognized by NEDD4L through a physical interaction. To address this, we first investigated the correlation between the AQP9 and NEDD4L protein level in CRC cells. NEDD4L overexpression resulted in an AQP9 protein level decrease (Figure 7B and C), while the AQP9 mRNA level was not affected (Figure 7D). Moreover, the half-life of AQP9 was reduced by NEDD4L (Figure 7E). Co-IP revealed the interaction between AQP9 and NEDD4L (Figure 7F). We then examined whether NEDD4L could ubiquitinate AQP9. Indeed, NEDD4L overexpression efficiently promoted AQP9 ubiquitination whereas knock-down of NEDD4L decreased AQP9 ubiquitination (Figure 7G and H). Consistently with the reported interaction between the WW domains of NEDD4L and the PY motifs of its substrates, mutation of the PY motif (LPFY to LAFA, AA mutant) of AQP9 abolished its interaction with NEDD4L and AQP9 (AA) could not be ubiquitinated (Figure 7I).
Figure 7.
The E3 ligase neural precursor cell expressed developmentally downregulated 4-like (NEDD4L) ubiquitinates and degrades AQP9 through the PY (LPFY) motif. (A) The C-terminal of AQP9 contains a PY motif. (B) and (C) Western blot analysis of AQP9 upon NEDD4L overexpression. (D) AQP9 mRNA level determined by using qPCR in HCT116 cells. (E) CHX chase assays to determine the half-life of AQP9 in NEDD4L-overexpressed HCT116 cells. (F) Co-IP reveals the interaction between AQP9 and NEDD4L. Representative immunoblots of an in vivo ubiquitination (Ub) assay of HEK293T cells transiently co-transfected with (G) pcDNA–his-Ub, GFP–AQP9 plasmids, and HA–NEDD4L plasmids or (H) siNEDD4L as well as their relative controls. (I) Co-IP experiment of Ub after being co-transfected with HA-NEDD4L plasmids and GFP–AQP9–WT plasmids or GFP–AQP9–AA (mutation of the PY motif (AA) of AQP9). All data are shown as mean ± SEM. *P < 0.05; **P < 0.01; ***P < 0.001. CHX, cycloheximide; Co-IP, co-immunoprecipitation; AQP9, aquaporin 9; WT, wild-type; SEM, standard error of the mean; ns, no significance.
Discussion
Substantial studies indicated roles for aquaporins in cancer development and metastasis [9–11, 37]. AQP1 deletion remarkably impaired cell migration, angiogenesis, and tumor growth [38]. AQP5 was found to contribute to gastric carcinogenesis [39]. In the aquaglyceroporin subfamily, AQP3 was associated with metastasis in a variety of cancers [40, 41]. AQP9 was known for regulating water and glycerol transport in hepatocytes, but has been implicated in liver cancer as a tumor suppressor [42]. We previously found that AQP9 was correlated with chemotherapy efficacy. However, the role and mechanism of AQP9 in CRC have not been fully elucidated. In the present study, we identified that AQP9 was highly expressed in metastatic CRC tumors. Higher AQP9 expression was significantly associated with advanced disease stage and unfavorable prognosis for patients with CRC and AQP9 overexpression promoted CRC metastasis in vitro and in vivo.
Recent study put forward an “Osmotic Engine Model,” elucidating that water permeation was a crucial mechanism of cell migration in the viscous extracellular matrix, where Na+/H+ exchanger 1 (NHE1) and AQP5 were involved [43, 44]. Interestingly, previous study also reported that AQP9 could form a complex with NHERF1 (Na+/H+ Exchanger Regulatory Factor), which was important in regulating AQP9 activation and glycerol-induced cell swelling [14]. Our observation showed that AQP9 overexpression could induce a mesenchymal morphology with increased cell motility. According to the osmotic model, overexpression of AQP9 in CRC cells probably triggered a series of intracellular processes including cell swelling through water and small molecule uptake, elevating cell membrane tension, etc. and eventually led to a protrusive cell morphology and faster migration. Further experiments need to be performed to elucidate the role of AQP9 in regulating CRC cell morphology change.
The association between aquaporins and the Wnt pathway had been reported by previous studies. Wnt5a regulated AQP2 expression, phosphorylation, and apical membrane localization in the kidney [45]. AQP5+ stem cells have been proven to be a source of Wnt-driven, invasive gastric cancer in vivo [39]. Our results revealed that AQP9 overexpression was associated with the Wnt pathway enrichment. Interestingly, we found that AQP9 was highly expressed in CMS4 CRC but not in canonical Wnt-associated CMS2 CRC. Study showed that physiological and oncogenic Wnt induced distinct prognostic responses. The Wnt-receptor signature was associated with poor prognosis and overlapped with the CMS4 CRC gene signatures [46]. Consistently, we found that AQP9 high expression was correlated with higher TNM stage and poorer prognosis in patients with CRC, suggesting that AQP9 might be associated with extrinsic Wnt-receptor activation.
DVL2 was a major PDZ-containing protein that facilitated protein complex formation, which was an essential step for the transduction of Wnt signals. Inhibitors targeting the PDZ-mediated interaction of DVL2 have been proven to be effective in blocking the Wnt pathway [8, 47]. As AQP9 possessed a putative PDZ-binding domain, we then determined whether AQP9 could bind to DVL2. Both artificial intelligence (AI) prediction and in vitro experiments confirmed that AQP9 bound to DVL2 via its SVIM motif. We also determined via the luciferase assay that deletion of the SVIM motif in AQP9 inhibited canonical Wnt signaling. Furthermore, high expression levels of AQP9 and DVL2 were significantly associated with low probability of OS and PFS in patients with CRC.
Given that DVL2 could recruit Axis inhibition protein 1 and other core components to the plasma membrane, then relayed Wnt signaling from receptors to β-catenin and downstream effectors [7], we further investigated the effects of AQP9 on DVL2 and β-catenin. We found that CRC exhibited parallel upregulation of AQP9, DVL2, and nuclear β-catenin. AQP9 overexpression significantly enhanced the half-life of DVL2 and β-catenin. The increased β-catenin stability is known to mainly contribute to the EMT phenotype. These findings, combined with the fact that AQP9 overexpression activated canonical Wnt signaling and CRC metastasis, provided strong evidence that AQP9 played an important role in regulating Wnt signaling.
The expression of AQP9 has been shown to be regulated by testosterone and estrogen. Nonetheless, the intracellular regulation mechanism is still not clear. It is well established that protein degradation by proteasome or lysosome via ubiquitination is a key regulator in numerous biological processes. We identified E3 ligase NEDD4L as an upstream regulator of AQP9. Moreover, our findings indicated that the PY motif might be critical for the ubiquitination and protein degradation of AQP9 modulated by NEDD4L.
Supplementary Material
Acknowledgements
Not applicable.
Contributor Information
Yiting Liu, Guangdong Institute of Gastroenterology, Guangdong Provincial Key Laboratory of Colorectal and Pelvic Floor Diseases, Guangzhou, Guangdong, P. R. China; Department of General Surgery (Colorectal Surgery), The Sixth Affiliated Hospital, Sun Yat-sen University, Guangzhou, Guangdong, P. R. China.
Qianling Gao, Guangdong Institute of Gastroenterology, Guangdong Provincial Key Laboratory of Colorectal and Pelvic Floor Diseases, Guangzhou, Guangdong, P. R. China; Department of General Surgery (Colorectal Surgery), The Sixth Affiliated Hospital, Sun Yat-sen University, Guangzhou, Guangdong, P. R. China.
Xingzhi Feng, Guangdong Institute of Gastroenterology, Guangdong Provincial Key Laboratory of Colorectal and Pelvic Floor Diseases, Guangzhou, Guangdong, P. R. China; Department of General Surgery (Colorectal Surgery), The Sixth Affiliated Hospital, Sun Yat-sen University, Guangzhou, Guangdong, P. R. China.
Guanxing Chen, Artificial Intelligence Medical Research Center, School of Intelligent Systems Engineering, Sun Yat-sen University, Shenzhen, Guangdong, P. R. China.
Xuefei Jiang, Guangdong Institute of Gastroenterology, Guangdong Provincial Key Laboratory of Colorectal and Pelvic Floor Diseases, Guangzhou, Guangdong, P. R. China; Department of General Surgery (Colorectal Surgery), The Sixth Affiliated Hospital, Sun Yat-sen University, Guangzhou, Guangdong, P. R. China.
Daici Chen, Guangdong Institute of Gastroenterology, Guangdong Provincial Key Laboratory of Colorectal and Pelvic Floor Diseases, Guangzhou, Guangdong, P. R. China; Department of General Surgery (Colorectal Surgery), The Sixth Affiliated Hospital, Sun Yat-sen University, Guangzhou, Guangdong, P. R. China.
Zihuan Yang, Guangdong Institute of Gastroenterology, Guangdong Provincial Key Laboratory of Colorectal and Pelvic Floor Diseases, Guangzhou, Guangdong, P. R. China; Department of Clinical Laboratory, The Sixth Affiliated Hospital, Sun Yat-sen University, Guangzhou, Guangdong, P. R. China.
Supplementary Data
Supplementary data is available at Gastroenterology Report online
Authors’ Contributions
Z.Y. conceived the study, designed and performed experiments, analysed data, and wrote the manuscript. Y.L. performed experiments, analysed data, and wrote the manuscript. Q.G. performed experiments, analysed data, and wrote the manuscript. X.F. performed experiments, analysed data, and wrote the manuscript. G.C. did artificial intelligence and machine-learning prediction analysis. X.J. analysed data. D.C. supervised. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the National Natural Science Foundation of China [No. 81702330 to Z.Y.] and Guangdong Natural Science Foundation [No. 2021A1515011400 to Z.Y.].
Conflict of Interest
The authors declare that there is no conflict of interests in this study.
References
- 1. Xie Y, Shi L, He X. et al. Gastrointestinal cancers in China, the USA, and Europe. Gastroenterol Rep (Oxf) 2021;9:91–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ciardiello F, Ciardiello D, Martini G. et al. Clinical management of metastatic colorectal cancer in the era of precision medicine. CA Cancer J Clin 2022;72:372–401. [DOI] [PubMed] [Google Scholar]
- 3. Siegel RL, Miller KD, Goding SA. et al. Colorectal cancer statistics, 2020. CA Cancer J Clin 2020;70:145–64. [DOI] [PubMed] [Google Scholar]
- 4. Biller LH, Schrag D.. Diagnosis and treatment of metastatic colorectal cancer: a review. JAMA 2021;325:669–85. [DOI] [PubMed] [Google Scholar]
- 5. Zhao H, Ming T, Tang S. et al. Wnt signaling in colorectal cancer: pathogenic role and therapeutic target. Mol Cancer 2022;21:144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Caspi M, Wittenstein A, Kazelnik M. et al. Therapeutic targeting of the oncogenic Wnt signaling pathway for treating colorectal cancer and other colonic disorders. Adv Drug Deliv Rev 2021;169:118–36. [DOI] [PubMed] [Google Scholar]
- 7. Gao C, Chen YG.. Dishevelled: the hub of Wnt signaling. Cell Signal 2010;22:717–27. [DOI] [PubMed] [Google Scholar]
- 8. Zhang Y, Appleton BA, Wiesmann C. et al. Inhibition of Wnt signaling by Dishevelled PDZ peptides. Nat Chem Biol 2009;5:217–9. [DOI] [PubMed] [Google Scholar]
- 9. De Ieso ML, Yool AJ.. Mechanisms of aquaporin-facilitated cancer invasion and metastasis. Front Chem 2018;6:135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Morishita K, Watanabe K, Ichijo H.. Cell volume regulation in cancer cell migration driven by osmotic water flow. Cancer Sci 2019;110:2337–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Login FH, Jensen HH, Pedersen GA. et al. Aquaporins differentially regulate cell-cell adhesion in MDCK cells. Faseb J 2019;33:6980–94. [DOI] [PubMed] [Google Scholar]
- 12. Huang D, Feng X, Liu Y. et al. AQP9-induced cell cycle arrest is associated with RAS activation and improves chemotherapy treatment efficacy in colorectal cancer. Cell Death Dis 2017;8:e2894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kreida S, Roche JV, Olsson C. et al. Protein-protein interactions in AQP regulation-biophysical characterization of AQP0-CaM and AQP2-LIP5 complex formation. Faraday Discuss 2018;209:35–54. [DOI] [PubMed] [Google Scholar]
- 14. Pietrement C, Da Silva N, Silberstein C. et al. Role of NHERF1, cystic fibrosis transmembrane conductance regulator, and cAMP in the regulation of aquaporin 9. J Biol Chem 2008;283:2986–96. [DOI] [PubMed] [Google Scholar]
- 15. Kim HY, Choi S, Yoon JH. et al. Small molecule inhibitors of the Dishevelled-CXXC5 interaction are new drug candidates for bone anabolic osteoporosis therapy. EMBO Mol Med 2016;8:375–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Consortium TU. UniProt: the universal protein knowledgebase. Nucleic Acids Res 2017;45:D158–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lu S, Wang J, Chitsaz F. et al. CDD/SPARCLE: the conserved domain database in 2020. Nucleic Acids Res 2020;48:D265–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Waterhouse A, Bertoni M, Bienert S. et al. SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res 2018;46:W296–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Yang J, Anishchenko I, Park H. et al. Improved protein structure prediction using predicted interresidue orientations. Proc Natl Acad Sci USA 2020;117:1496–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zhang Y. I-TASSER server for protein 3D structure prediction. BMC Bioinformatics 2008;9:40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Chen VB, Arendall WB 3rd, Headd JJ. et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr 2010;66:12–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Spassov VZ, Flook PK, Lisa Y.. LOOPER: a molecular mechanics-based algorithm for protein loop prediction. Protein Eng Des Sel 2008;21:91–100. [DOI] [PubMed] [Google Scholar]
- 23. Dienstmann R, Vermeulen L, Guinney J. et al. Consensus molecular subtypes and the evolution of precision medicine in colorectal cancer. Nat Rev Cancer 2017;17:79–92. [DOI] [PubMed] [Google Scholar]
- 24. Guinney J, Dienstmann R, Wang X. et al. The consensus molecular subtypes of colorectal cancer. Nat Med 2015;21:1350–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wu ZQ, Brabletz T, Fearon E. et al. Canonical Wnt suppressor, Axin2, promotes colon carcinoma oncogenic activity. Proc Natl Acad Sci USA 2012;109:11312–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wong HC, Bourdelas A, Krauss A. et al. Direct binding of the PDZ domain of Dishevelled to a conserved internal sequence in the C-terminal region of Frizzled. Mol Cell 2003;12:1251–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kaur P, Lam VYM, Mannava AG. et al. Membrane targeting of disheveled can bypass the need for arrow/LRP5. Sci Rep 2017;7:6934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kaeser PS, Deng L, Wang Y. et al. RIM proteins tether Ca2+ channels to presynaptic active zones via a direct PDZ-domain interaction. Cell 2011;144:282–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kim E, Niethammer M, Rothschild A. et al. Clustering of Shaker-type K+ channels by interaction with a family of membrane-associated guanylate kinases. Nature 1995;378:85–8. [DOI] [PubMed] [Google Scholar]
- 30. Kastritis PL, Bonvin AM.. On the binding affinity of macromolecular interactions: daring to ask why proteins interact. J R Soc Interface 2013;10:20120835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ozdal T, Capanoglu E, Altay F.. A review on protein–phenolic interactions and associated changes. Food Res Int 2013;51:954–70. [Google Scholar]
- 32. Goel P, Manning JA, Kumar S.. NEDD4-2 (NEDD4L): the ubiquitin ligase for multiple membrane proteins. Gene 2015;557:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Persaud A, Jiang C, Liu Z. et al. Elevated intracellular Na(+) and osmolarity stimulate catalytic activity of the ubiquitin ligase NEDD4-2. Proc Natl Acad Sci USA 2022;119:e2122495119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Duerr J, Leitz DHW, Szczygiel M. et al. Conditional deletion of NEDD4-2 in lung epithelial cells causes progressive pulmonary fibrosis in adult mice. Nat Commun 2020;11:2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Jiang C, Kawabe H, Rotin D.. The ubiquitin ligase NEDD4L regulates the Na/K/2Cl co-transporter NKCC1/SLC12A2 in the colon. J Biol Chem 2017;292:3137–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wang C, An J, Zhang P. et al. The NEDD4-like ubiquitin E3 ligases target angiomotin/p130 to ubiquitin-dependent degradation. Biochem J 2012;444:279–89. [DOI] [PubMed] [Google Scholar]
- 37. Marlar S, Jensen HH, Login FH. et al. Aquaporin-3 in cancer. IJMS 2017;18:2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Saadoun S, Papadopoulos MC, Hara-Chikuma M. et al. Impairment of angiogenesis and cell migration by targeted aquaporin-1 gene disruption. Nature 2005;434:786–92. [DOI] [PubMed] [Google Scholar]
- 39. Tan SH, Swathi Y, Tan S. et al. AQP5 enriches for stem cells and cancer origins in the distal stomach. Nature 2020;578:437–43. [DOI] [PubMed] [Google Scholar]
- 40. Xiong G, Chen X, Zhang Q. et al. RNA interference influenced the proliferation and invasion of XWLC-05 lung cancer cells through inhibiting aquaporin 3. Biochem Biophys Res Commun 2017;485:627–34. [DOI] [PubMed] [Google Scholar]
- 41. Chen J, Wang T, Zhou YC. et al. Aquaporin 3 promotes epithelial-mesenchymal transition in gastric cancer. J Exp Clin Cancer Res 2014;33:38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. da Silva IV, Garra S, Calamita G. et al. The multifaceted role of aquaporin-9 in health and its potential as a clinical biomarker. Biomolecules 2022;12:897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Bera K, Kiepas A, Godet I. et al. Extracellular fluid viscosity enhances cell migration and cancer dissemination. Nature 2022;611:365–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Stroka KM, Jiang H, Chen SH. et al. Water permeation drives tumor cell migration in confined microenvironments. Cell 2014;157:611–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ando F, Sohara E, Morimoto T. et al. Wnt5a induces renal AQP2 expression by activating calcineurin signalling pathway. Nat Commun 2016;7:13636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Michels BE, Mosa MH, Grebbin BM. et al. Human colon organoids reveal distinct physiologic and oncogenic Wnt responses. J Exp Med 2019;216:704–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Spassov VZ, Yan L.. A fast and accurate computational approach to protein ionization. Protein Sci 2008;17:1955–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analysed in this study are included in this published article. All the data supporting the findings of this study are available from the corresponding author upon request.