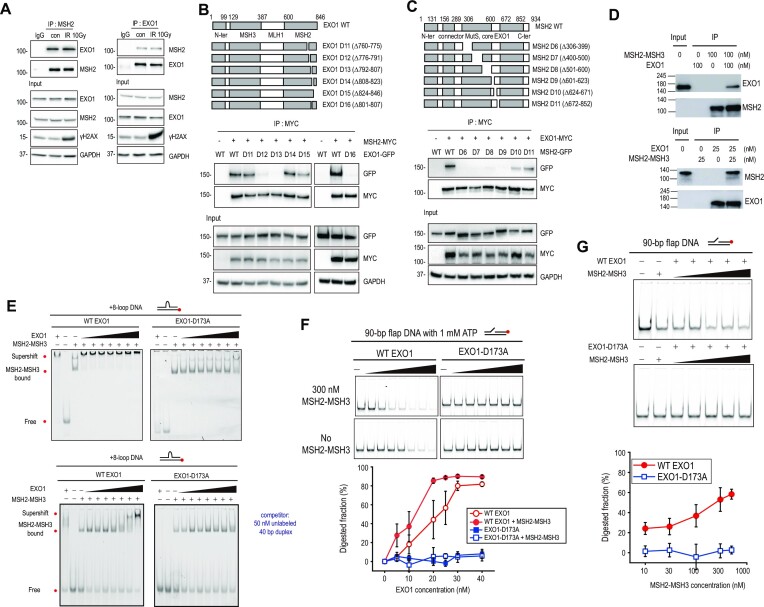
Figure 3.
MSH2 interaction with EXO1 enhances EXO1 activity. (A) For endogenous immunoprecipitation, HEK293T cells were not irradiated or irradiated with 10 Gy of ionizing radiation. Cell extracts were incubated with anti-IgG, anti-MSH2, or anti-EXO1 antibody. Proteins immunoprecipitated with Dynabeads Protein G were analyzed by western blot. For 10 Gy exposure, γH2AX was used as a DNA damage marker. (B) Diagram of EXO1 WT and EXO1 deletion mutants. HEK293T cells were co-transfected with myc-MSH2 WT and GFP-EXO1 or GFP-EXO1 deletion mutants. Interaction of each EXO1 deletion mutant with MSH2 was determined by immunoprecipitation with MYC antibody. (C) Diagram of MSH2 WT and MSH2 deletion mutants. HEK293T cells were co-transfected with myc-EXO1 WT and each GFP-MSH2 deletion mutant. (D) Immunoprecipitation assays for purified EXO1 and MSH2-MSH3 proteins. Indicated antibodies were used for western blotting. (E) EMSA for MSH2-MSH3 and EXO1. MSH2-MSH3 (100 nM) was bound to +8-loop DNA, and WT EXO1 (left) or EXO1 nuclease mutant (Mut EXO1-D173A) (right), and in the absence (top) or presence (bottom) of competitor was titrated (0, 10, 15, 20, 25, 30, 40, and 80 nM). (F) Nuclease activity of EXO1 in the presence or absence of MSH2-MSH3 with 1 mM ATP. DNA (40 nM) with 90 bp flap DNA was reacted with WT EXO1 or Mut EXO1-D173A at different concentrations (0, 5, 10, 20, 25, 30, and 40 nM) in the presence (top) or absence (bottom) of 300 nM MSH2-MSH3. Quantification is shown below the gel images. Error bars represent standard error determined from triplicate samples. (G) Nuclease activity of EXO1 in the titration of MSH2-MSH3. 40 nM DNA with 90 bp flap DNA was reacted with 20 nM WT EXO1 (top) or Mut EXO1-D173A (middle) at different concentrations (0, 10, 30, 100, 300, and 500 nM) of MSH2-MSH3. The EXO1 nuclease activity was quantified (bottom). Error bars were obtained from standard error in triplicate.