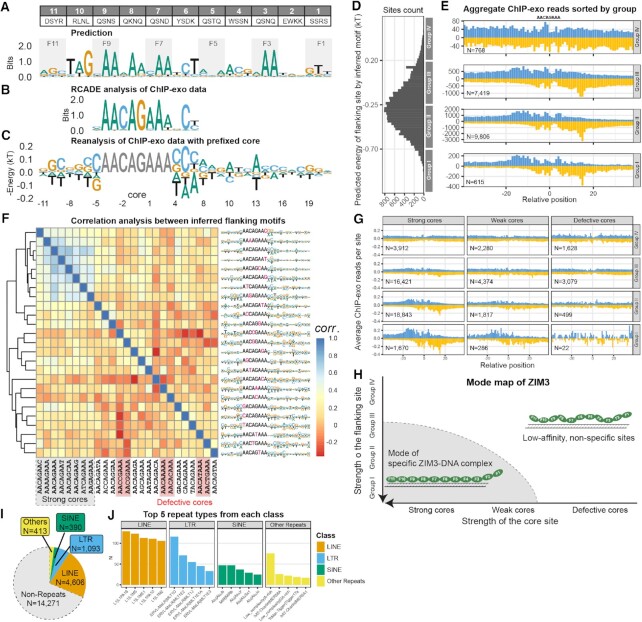
Figure 5.
ModeMap analysis of ZIM3 reveals its extended motif, recognition model, and bound repeat elements. (A) Contact residues for human ZIM3; Motif prediction by B1H method. (B) Motif from RCADE analysis of ChIP-exo data. (C) Extended motif by reanalysis of ChIP-exo data with prefixed core AACAGAAA. (D) Distribution of binding sites according to the predicted energy of flanking sequences. (E) Aggregate ChIP-exo reads distribution sorted by groups with equal energy bandwidth. (F) Extended motifs by reanalysis of ChIP-exo data with all single variants of AACAGAAA as the prefixed core; Heatmap is generated by auto-correlation analysis of all extended motifs; Strong and defective cores are labeled with grey and red shaded boxes; remaining cores are classified as weak cores. (G) Re-plot of aggregate ChIP-exo signals around three classes of cores, each sorted by the flanking site strength as E) and normalized by the number of sites. (H) Mode map inferred from G) (I) Repeats distribution of in vivo specific sites identified within the full mode inferred from ModeMap. (J) Top 5 repeat types from each class among identified sites.