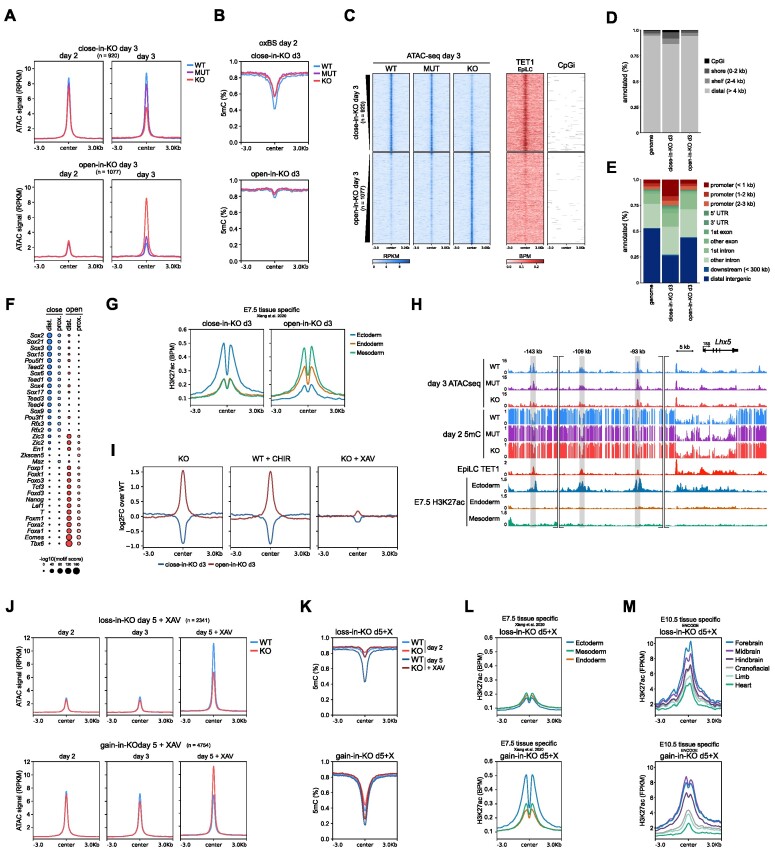
Figure 5.
Non-catalytical regulation of chromatin accessibility by TET1 at distal enhancers. (A) Profiles of ATAC-seq signal in reads per kilobase million (RPKM) centred at day 3 close-in-KO (top panels) or open-in-KO (bottom panels) differentially accessible regions (DARs) at day 2 (left) and day 3 (right) of neurobasal differentiation in Tet1 KO, MUT and WT cells. (B) Profiles of 5mC levels at day 2, centred at day 3 DARs. (C) Heatmaps of ATAC-seq signal in RPKM in KO, MUT and WT cells at day 3 of differentiation, TET1 binding in BPM in EpiLCs, and CpGi frequency centred at day 3 close-in-KO (top panels) and open-in-KO (bottom panels) DARs ordered by fold change of differential accessibility. (D, E) Genomic features based on CpG island proximity (D) and gene structure (E) annotation of close-in-KO and open-in-KO DARs at day 3. (F) Scores for top 20 close-in-KO and open-in-KO transcription factor (TF) motifs in distal (close-in-KO d3, n = 625, 68%; open-in-KO d3, n = 902, 84%) or proximal (close-in-KO d3, n = 295, 32%; open-in-KO d3, n = 175, 16%) DARs, filtered by a minimum TF expression of 5 TPM on at least one time-point during the RNA-seq time-course. A DAR was considered to be distal if the distance to TSS was >3 kb. (G) Profiles of H3K27ac ChIP-seq in BPM in E7.5 germ layer-specific tissues (Xiang et al., 2020) centred at day 3 close-in-KO and open-in-KO DARs (46). (H) IGV tracks for ATAC-seq signal at day 3, 5mC levels at day 2, TET1 binding in EpiLCs and E7.5 germ layer-specific H3K27ac ChIP-seq signal at three Lhx5 distal enhancers. Close-in-KO DARs are highlighted in grey. (I) Log2 fold changes of ATAC-seq signal in KO cells at day 3, WT cells treated for 24 h with 3 μM of the Wnt activator CHIR99021 (CHIR), and KO cells treated for 24 h with the Wnt inhibitor XAV939 (XAV), compared to day 3 WT cells, centred at day 3 close-in-KO and open-in-KO DARs. (J) Profiles of ATAC-seq signal in RPKM centred at day 5 (+XAV) loss-in-KO (top panels) or gain-in-KO (bottom panels) differentially accessible regions (DARs) at day 2 (left), day 3 (middle), and day 5 (+XAV) (right) of differentiation in Tet1 KO and WT cells. (K) Profiles of true 5mC levels at day 2 and day 5 (+XAV), centred at day 5 (+XAV) DARs. (L) Profiles of H3K27ac ChIP-seq in BPM in E7.5 germ layer-specific tissues (Xiang et al., 2020) centred at day 5 (+XAV) loss-in-KO and gain-in-KO DARs (46). (M) Profiles of H3K27ac ChIP-seq in FPKM in ENCODE E10.5 mouse tissues (31), centred at day 5 (+XAV) DARs.