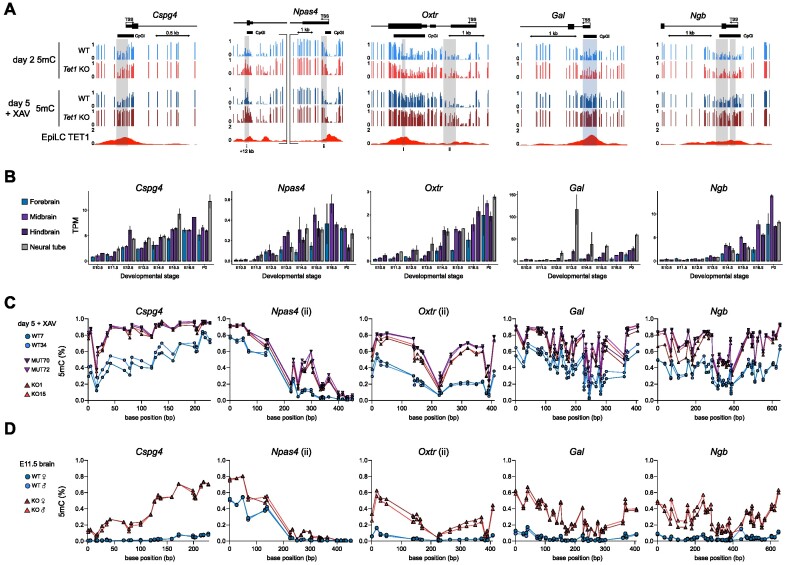
Figure 7.
Early developmental origin of disease-associated DMRs in Tet1−/− mice. (A) IGV tracks of 5mC methylation levels in KO and WT cells at day 2 and day 5 (+XAV), and TET1 occupancy in EpiLCs, at 5 genes known to be hypermethylated in adult Tet1 KO mice and implicated in neural dysfunction: Cspg4, Gal, and Ngb (Cheng et al.), Npas4 (Rudenko et al.) and Oxtr (Williams et al.). DMRs are highlighted grey. At Gal, a region of interest bound by TET1, though not a DMR, is highlighted in blue. (B) RNA-seq expression in TPM of indicated genes in specific tissues per developmental stage from ENCODE (31), shown as mean ± SEM of n = 2 samples per condition. (C, D) 5mC levels at hyper DMRs at the promoters of Cspg4, Gal, Ngb, Npas4 (region ii), and Oxtr (region ii) in Tet1 KO, MUT and WT (C) day 5 (+XAV) antNPCs and (D) E11.5 brains from sex and stage-matched littermates, measured using amplicon bisulfite sequencing.