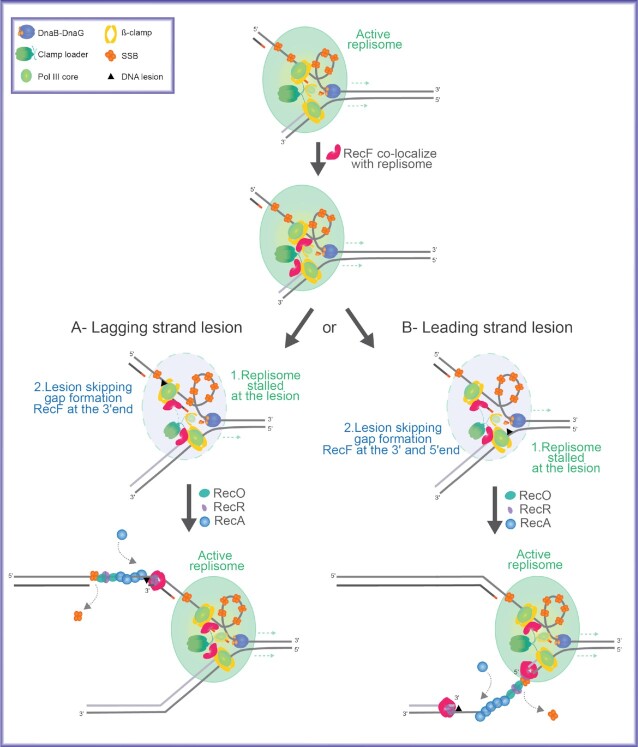
Figure 10.
Model of RecF ATPase function near the replisome. Schematic representation of RecF ATPase activity triggering the localization of RecF near the replisome. The light green circle represents the stable replisome. In the front of the replication fork the DnaB helicase (dark blue) unwinds the dsDNA and interacts with DnaG (dark orange). DnaG promotes RNA priming on the lagging strand. On the lagging strand, the ssDNA region intermittently formed during replication is coated by the SSB protein (light orange tetramer). The clamp loader (dark green) interacts with the two polymerase cores of the leading and lagging strand, the clamp loader also interacts with a third PolIII core that would be loaded on the next RNA primed site. Those interactions allow the integrity of the replisome. The β-clamp (yellow) increases the processivity of PolIII is represented in yellow. Gap formation occurs upon encounter with a lesion. RecF (pink), initially associated with the replisome, is deposited at the 3′ end of the interrupted DNA strand. Stability of the bound RecF is increased by binding to RecR (purple). Gaps formed on the lagging strand have RecFR at one end of the gap. Gaps formed in the leading strand may have RecFR positioned at one or both ends of the gap. Finally, RecA is loaded onto the SSB coated DNA by the RecO (marine green) and RecR proteins at a site within the gap, potentially facilitated by RecR handoff from RecFR. The authors encourage readers to compare the elements of this model with speculation offered by Kuzminov in 1999 (60).