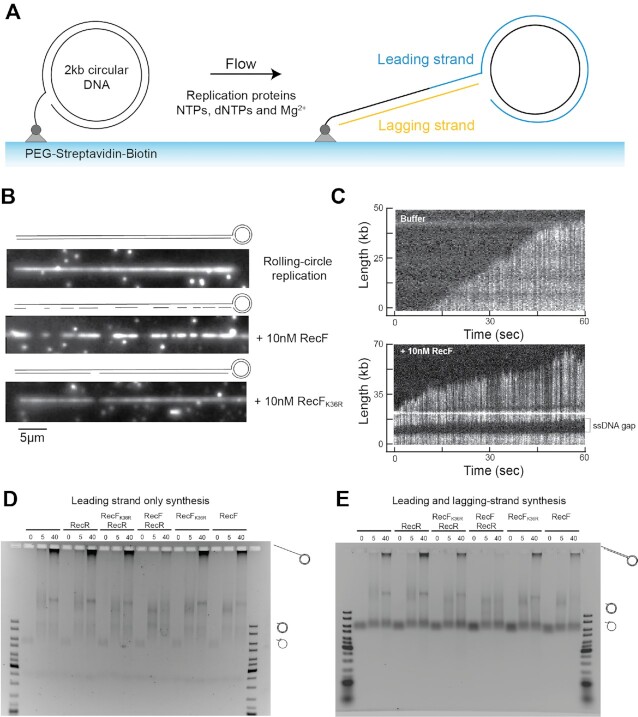
Figure 9.
Gap formation during in vitro replication at physiological concentrations of RecF ATPase protein. The capacity of physiological concentrations of RecF ATPase protein to create DNA gaps during replication was tested in vitro. (A) Schematic representation of the experimental design. Circular 5′-biotinylated DNA is coupled to the functionalized surface of a microfluidic flow cell through a streptavidin linkage. Addition of E. coli replication proteins and nucleotides results in the initiation of DNA synthesis. Newly synthetized DNA products are extended by flow, labelled with DNA stain and visualized in real time using fluorescence microscopy. (B) Example of individual DNA molecules produced during pre-assembled rolling-circle replication in the absence of RecF, or in presence of 10 nM RecF or RecFK36R proteins. The gray scale indicates the fluorescence intensity of stained DNA. (C) Kymographs showing the progression of DNA synthesis during rolling-circle replication without or with the addition of 10 nM RecF. A gap is identified by discontinuity of fluorescence intensity in a DNA molecule (D). Replication assay realized in batch on primed m13 circular DNA in the absence of the primase and the ribonucleotides, allowing only the replication of the leading strand. Samples of the ongoing replication were stopped at the indicated time. As mentioned, 20 nM of RecF or RecFK36R or 40 nM RecR protein are added. (E). Replication assays realized in batch on primed M13 circular DNA including primase and the ribonucleotides, allowing the replication of both leading and lagging strands. Proteins are added at the same concentration as the batch replication of the leading strand only.