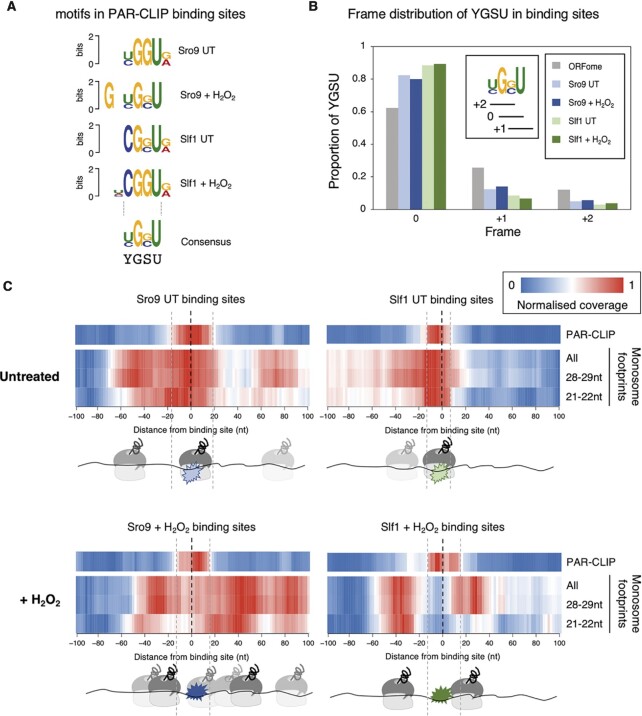
Figure 5.
LARP binding sites align with ribosome footprints. (A) The most enriched motif identified within PAR-CLIP mRNA binding sites in each condition, using DREME, aligned around the common GSU triplet. (Motif E-values (top to bottom): 1.1 × 10−98, 6.0 × 10−153,1.2 × 10−30, 4.5 × 10−16). (B) Reading frame distribution of GSU framing within PAR-CLIP binding sites and within ORFome, colouring as figure 1B. Inset shows the 0, + 1 and + 2 framing with respect to YGSU. (C) Ribosome read density around PAR-CLIP binding site mode locations (Supplementary Table S3) (-100 upstream to + 100 nt downstream). Plots show normalised sequence read coverage (enriched red) from either UT (top) or stressed (+ H2O2, bottom) conditions. Aligned PAR-CLIP reads for Sro9 (left) and Slf1 (right) cluster around 0. 80S monosome ribosome reads from a recent published study (4) mapping within each 200 nt binding site window are shown below each plot for standard ribosome footprints (28–29 nts) or small rotated ribosome footprints (21–22 nts) representing rotated ribosomes with free A-sites and both of these sets combined (All). Cartoons below each plot indicate relative positions of each LARP and the apparent preferred ribosome positions.