Fig. 5 |. Tumours generate ATP slower than healthy tissues.
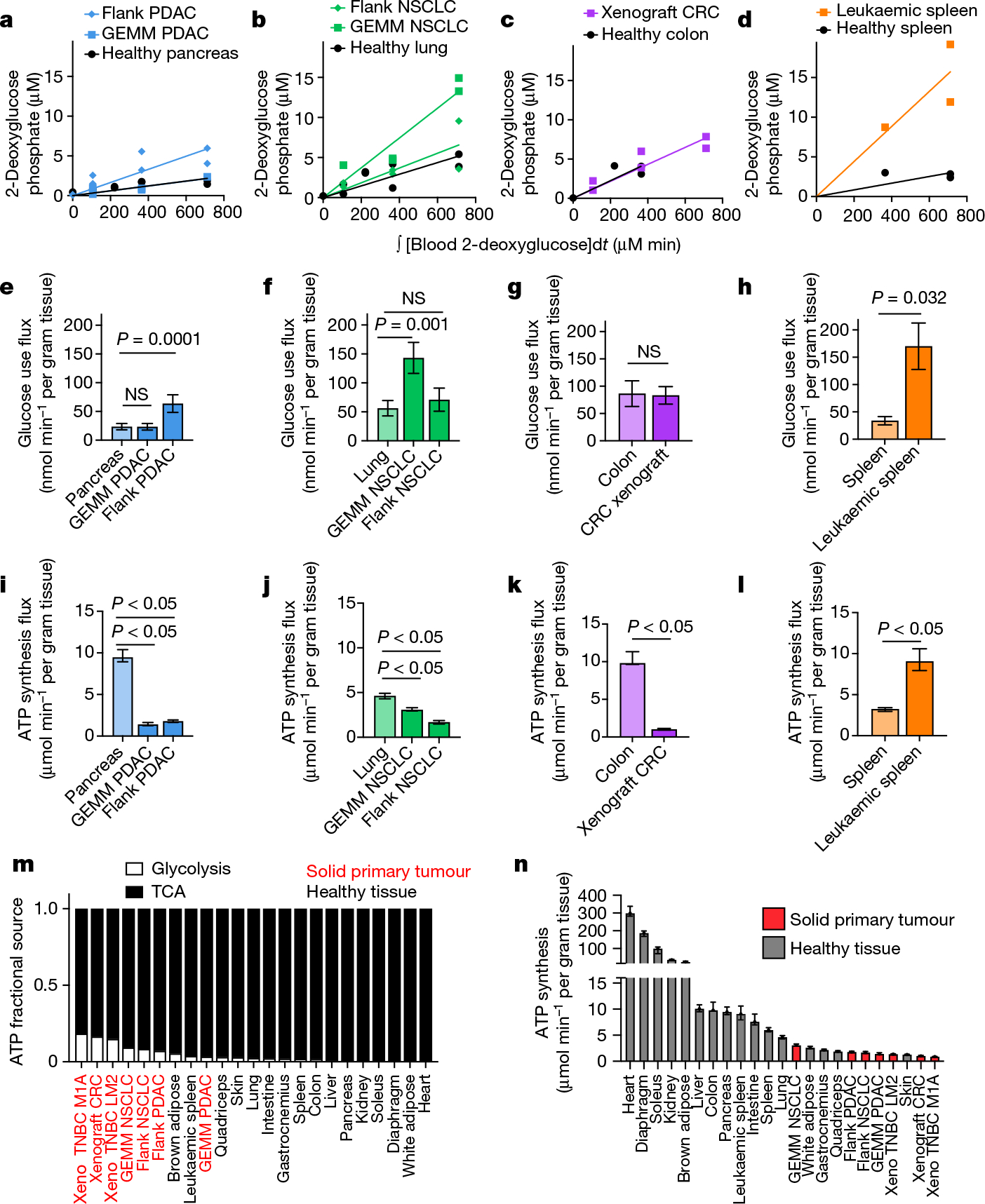
a–h, Quantification of tumour glucose use flux. Tissue [1-13C]2-deoxyglucose phosphate concentration versus the integral of blood [1-13C]2-deoxyglucose concentration with respect to time in healthy pancreas, GEMM PDAC and flank PDAC (n = 9, 5 and 6 mice, respectively) (a), in healthy lung, GEMM NSCLC and flank NSCLC (n = 9, 5 and 6 mice, respectively) (b), in healthy colon and xenograft CRC (n = 4 and 6 mice, respectively) (c) and in healthy spleen and leukaemic spleen (n = 3 mice each) (d). e–h, The glucose use flux in healthy tissues and cancer models calculated from a–d (pancreas (e), lung (f), colon (g) and spleen (h)). n values are shown. Data are value determined by linear fit ± s.d. i–l, The calculated total ATP production flux in healthy tissues and cancer models (pancreas (i), lung (j), colon (k) and spleen (l)), calculated from the glucose use fluxes shown in e–h and TCA fluxes shown in Fig. 3b–h. Glucose use fluxes: n = 9 mice (pancreas), n = 5 (GEMM PDAC), n = 6 (flank PDAC), n = 9 (lung), n = 5 (GEMM NSCLC), n = 6 (flank NSCLC), n = 4 (colon), n = 6 (xenograft CRC), n = 3 (spleen), n = 3 (leukaemic spleen); lactate infusion timepoints: n = 12 mice (pancreas), n = 6 (GEMM PDAC), n = 13 (flank PDAC), n = 12 (lungs), n = 9 (GEMM NSCLC), n = 10 (flank NSCLC), n = 12 (colon), n = 7 (xenograft CRC), n = 12 (spleen), n = 15 (leukaemic spleen); tissue metabolite concentrations: n = 4 mice for all except n = 3 for healthy spleen. Data are best estimates of flux from computational fitting of experimental data ± s.d. m, The fraction of total ATP derived from glycolysis or the TCA cycle in tumours and healthy tissues. n, The calculated total ATP production flux in healthy tissues and cancer models. Data are best estimates of flux from computational fitting of experimental data ± s.d. All healthy tissues in this figure were from non-tumour-bearing mice. For e–l, P values were calculated using two-tailed t-tests; NS, not significant. Tumour name abbreviations are the same as in Fig. 3, with the addition of MDA-MB-231-LM2 triple-negative breast cancer xenograft tumour (xeno TNBC LM2) and Sum159-M1A triple-negative breast cancer xenograft tumour (xeno TNBC M1A).
