Figure 1.
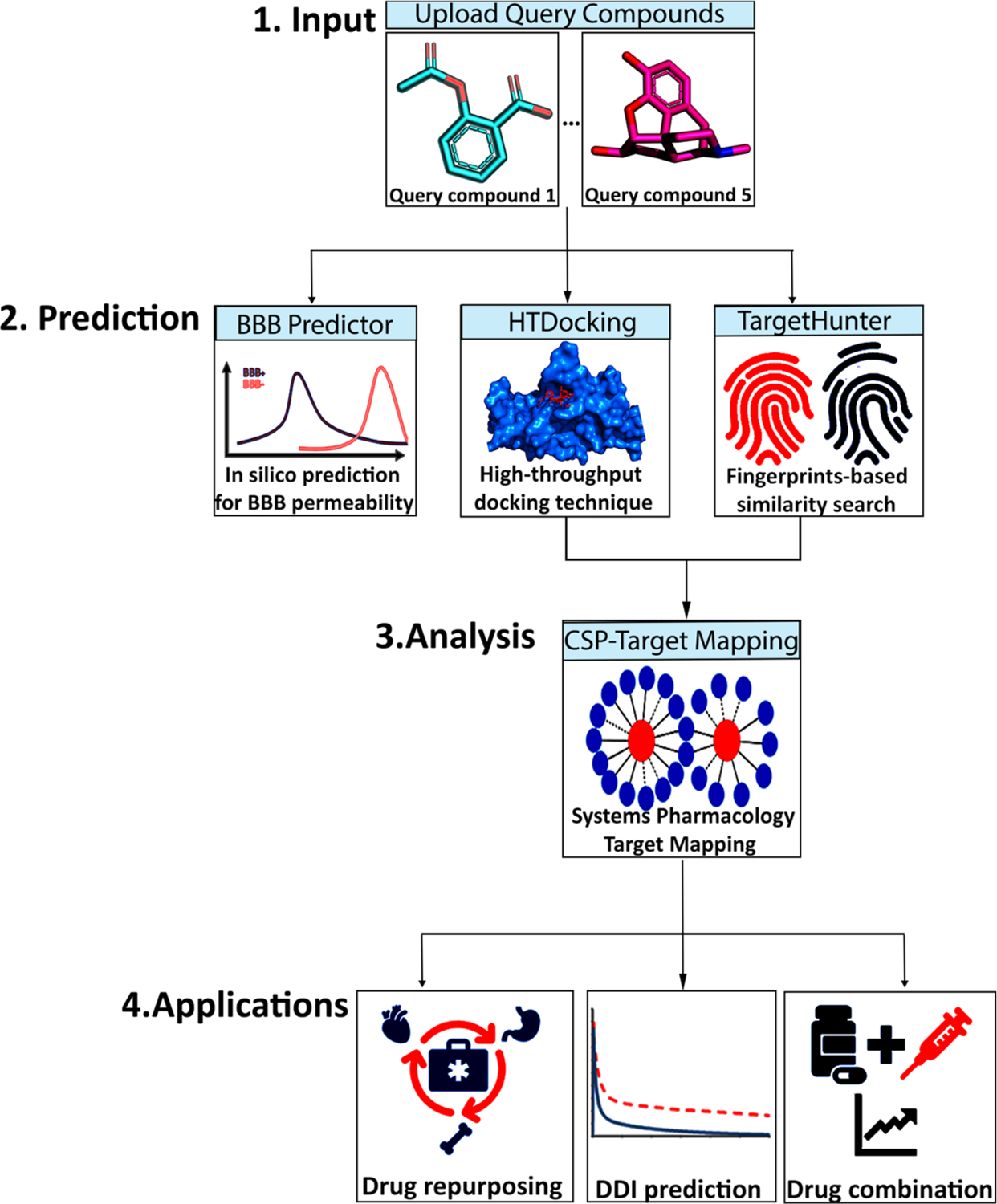
Workflow of the Pain-CKB server that is divided into three major steps: (i) input of chemical agent; (ii) in silico BBB prediction, high-throughput docking (HTDocking) with pain-related targets, and fingerprints-based similarity search (TargetHunter) by our established algorithms implemented in Pain-CKB; (iii) systems pharmacology target mapping for potential drug repurposing, DDI prediction, and drug combination.
