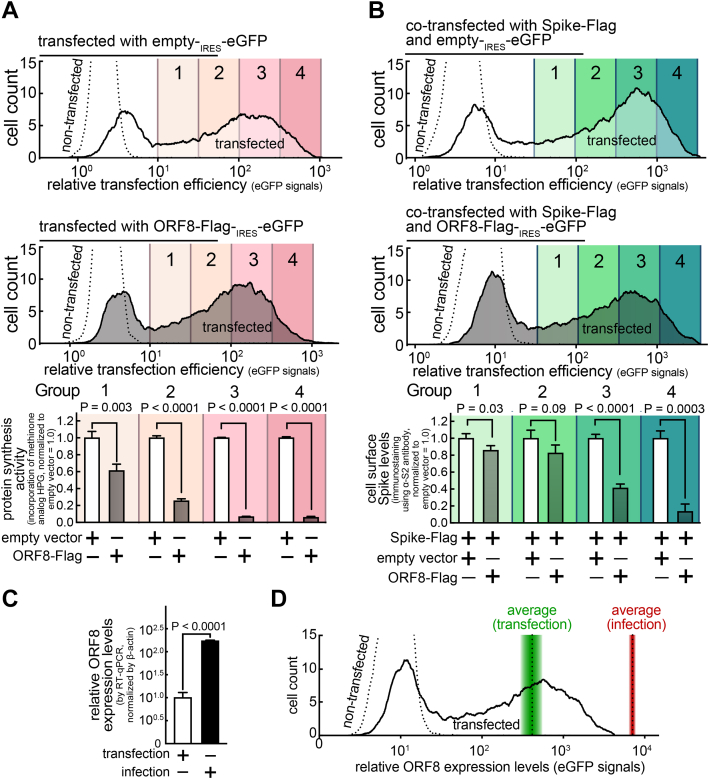
Figure 6.
The ORF8 cellular effects are dependent on the ORF8 expression levels, which were detected lower than ORF8 expression levels in SARS-CoV-2 infection.A and B, a monolayer of HEK293T cells were cotransfected with a bicistronic eGFP plasmid encoding none (top) or ORF8-Flag (middle) without (A) or with (B) a plasmid encoding Spike-Flag. After 30 min incubation in the absence (B) or presence (A) of HPG (methionine analog that can be fluorescently labeled), the cells were fixed, permeabilized, and fluorescently labeled for incorporated HPG (A) or immunostained for cell surface Spike levels using antibodies against Spike S2 (B). The cells were analyzed by flow cytometry and gated into four groups based on the transfection levels (eGFP) (100.5 fold increment). C and D, HEK293T cells stably expressing ACE2/TMPRSS2 were transfected with a bicistronic eGFP plasmid encoding ORF8-Strep for 18 h or infected with SARS-CoV-2 at the MOI of 0.1 for 48 h. The cells were lysed and analyzed by RT-qPCR using primers targeting ORF8 (C) or directly evaluated by flow cytometry analysis for relative ORF8 expression levels (eGFP signals) (D). The average eGFP signals of the whole transfected population is indicated (dotted line within green gradient), which was overlayed by the total ORF8 expression levels in the whole cell populations detected in C (gradient indicates s.d.). The data represent or are combined from three independent experiments and are presented as mean ± s.d. Statistical significance was analyzed using two-tailed Student’s t test. HPG, homopropargylglycine; RT-qPCR, reverse transcription-quantitative polymerase chain reaction; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2.