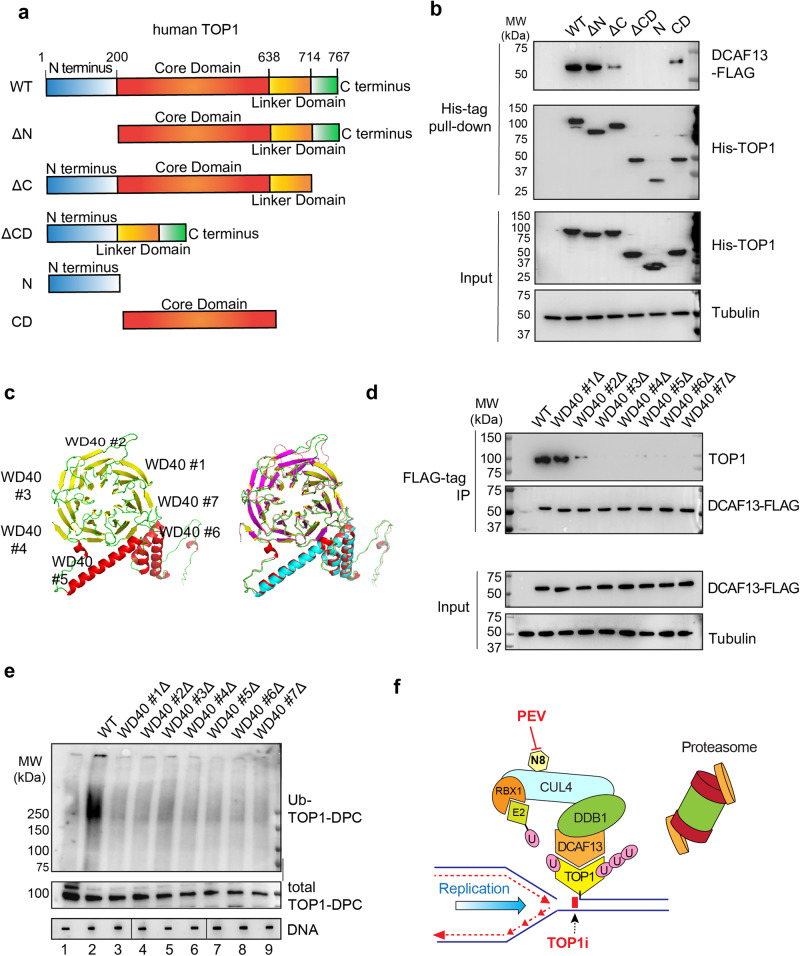
Fig. 7. DCAF13 interacts with the core domain of TOP1 using its putative WD40 domains.
a Domain schematics of human TOP1. b HCT116 cells were co-transfected with DCAF13-FLAG overexpression plasmid and indicated 6×His TOP1 constructs, followed by CPT (20 μM, 30 min) in the presence of BTZ (1 μM) for His-tag pull-down. The pull-down samples and cellular lysates (input) were subjected to IB using indicated antibodies. c Model structure of human DCAF13 predicted by AlphaFold248 (yellow, green, red) superimposed with SOF1 (yeast ortholog of human DCAF13 PDB: 6ZQB_46, red, magenta, blue). d HCT116 cells were transfected with the indicated DCAF13-FLAG constructs, followed by CPT (20 μM, 30 min) in presence of BTZ (1 μM) for FLAG-IP. The IP samples and cellular lysates (input) were subjected to IB using indicated antibodies. e HCT116 cells were transfected with the indicated DCAF13-FLAG constructs followed by CPT (20 μM, 30 min) for DUST assay for immunodetection of ubiquitylated TOP1-DPC and total TOP1-DPCs using anti-ubiquitin and anti-TOP1 antibodies. The order of DNA slot blots has been altered, as indicated by the line, and that uncropped labeled blots can be found in the Source Data file. N = 1. f TOP1-DPC arrest the replication forks upon their collision and signal the DCAF13-DDB1-CUL4-RBX1 complex for its recruitment to the DPCs. DCAF13 the substrate receptor that binds TOP1-DPCs and links the DPCs to the CRL4 complexes. CUL4 is activated by mono-neddylation by RBX1, which facilitates the transfer of ubiquitin from E2 and RBX1 to TOP1-DPC. The ubiquitylation leads to proteasomal degradation of TOP1-DPCs at arrested replication forks, leading to exposure of the otherwise concealed seDSB. N8 NEDD8, U ubiquitin.